For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LHVLRPGYAGAVNGHSNNGNDDETSTTPTLYIFPHAGGDATYYVPFSREFSADIKRIAVHYPGQRDGYGL PALTSIPALADEIFAIMKPSAPPEGAVAFFGHSMGGMLAFEVALRFQSAGYRLIALFVSACSAPGYIRYK QIKDFSDNDMLDLVVRMTGMNPDFFEDEEFRVGVLPTLRAARIIAGYNCPPETTVSCPIYTYIGDKDWIA TQEDMKPWRERTTGAFAIRVFPGDHFYLNGNLSELVCDIEDKTLEWCDRA*
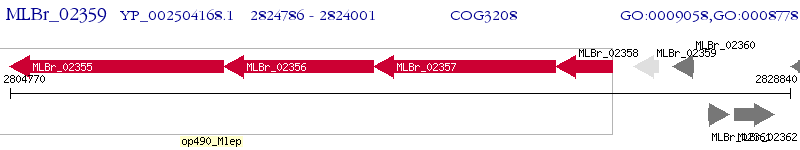
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. leprae Br4923 | MLBr_02359 | - | - | 100% (261) | thioesterase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2953 | tesA | 1e-112 | 74.12% (255) | thioesterase TESA |
| M. gilvum PYR-GCK | Mflv_3397 | - | 5e-62 | 50.42% (236) | thioesterase |
| M. tuberculosis H37Rv | Rv2928 | tesA | 1e-112 | 74.12% (255) | thioesterase TESA |
| M. abscessus ATCC 19977 | MAB_2033 | - | 2e-27 | 33.80% (216) | thioesterase |
| M. marinum M | MMAR_1778 | tesA | 1e-106 | 71.77% (248) | thioesterase TesA |
| M. avium 104 | MAV_2010 | - | 2e-20 | 28.64% (220) | phenyloxazoline synthase mbtb |
| M. smegmatis MC2 155 | MSMEG_4514 | - | 4e-24 | 29.61% (233) | thioesterase domain-containing protein |
| M. thermoresistible (build 8) | TH_4495 | - | 7e-24 | 31.96% (219) | PUTATIVE Thioesterase |
| M. ulcerans Agy99 | MUL_2025 | tesA | 1e-106 | 71.77% (248) | thioesterase TesA |
| M. vanbaalenii PYR-1 | Mvan_3130 | - | 1e-68 | 51.85% (243) | thioesterase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_4514|M.smegmatis_MC2_155 -----MTGTEVSLESWIKRYPAENTASG-PTLVFPHAGGAALAYRPFALA
TH_4495|M.thermoresistible__bu -----MTVLRQDFAPWIKRFPAGGERTGRAAVLFPHAGGAAAAYRTLAKA
MAV_2010|M.avium_104 --------------------------------MFPHAGAAAASYRVLAAA
MAB_2033|M.abscessus_ATCC_1997 ---MRRTDELLPGNPWLRRLRDGNGATR--VICFPHAGGSASYFRRLAKM
Mb2953|M.bovis_AF2122/97 MLARHGPRYGGSVNGHSDDSSGDAKQAAPTLYIFPHAGGTAKDYVAFSRE
Rv2928|M.tuberculosis_H37Rv MLARHGPRYGGSVNGHSDDSSGDAKQAAPTLYIFPHAGGTAKDYVAFSRE
MLBr_02359|M.leprae_Br4923 MLHVLRPGYAGAVNGHSNNGNDDETSTTPTLYIFPHAGGDATYYVPFSRE
MMAR_1778|M.marinum_M ------------MNGRSSNSKSDEKLTAPTLYIFPHAGGTAKDYVPFAKE
MUL_2025|M.ulcerans_Agy99 ------------MNGRSSNSKSDEKLTAPTLYIFPHAGGTAKDYVPFAKE
Mflv_3397|M.gilvum_PYR-GCK ----------------MKGEAG----VQPKLFIFPHAGGTPQYYVPFAKT
Mvan_3130|M.vanbaalenii_PYR-1 ---------------MTTGGAGESSAGQPKLFIFPHAGGSAQYYVPFAKT
*****. . : ::
MSMEG_4514|M.smegmatis_MC2_155 LAEAGSDAYVMQYPRRGDRLSHPAPATVGDLARDLFEAGDWTRLG-----
TH_4495|M.thermoresistible__bu LSDNGTDTYIVQYPQRAERLRHPAADSISDLAAQLFEAGPWGSVG-----
MAV_2010|M.avium_104 LA-AGADTYVVQYPQRAERLGDPAHESVHDLAVGLFRAAPWPGVA-----
MAB_2033|M.abscessus_ATCC_1997 LP-ADTEVLAVQYPGRGNRLREQPIASVPLLAEGIASALPASDK------
Mb2953|M.bovis_AF2122/97 FS-ADVKRIAVQYPGQHDRSGLPPLESIPTLADEIFAMMKPSAR---IDD
Rv2928|M.tuberculosis_H37Rv FS-ADVKRIAVQYPGQHDRSGLPPLESIPTLADEIFAMMKPSAR---IDD
MLBr_02359|M.leprae_Br4923 FS-ADIKRIAVHYPGQRDGYGLPALTSIPALADEIFAIMKPSAP---PEG
MMAR_1778|M.marinum_M FS-GEVKRVAVQYPGQQDGYGLPPLESIPGLAEEIFAIMKPAAR---IDT
MUL_2025|M.ulcerans_Agy99 FS-GEVKRVAVQYPGQQDGYGLPPLESIPGLAEEIFAIMKPAAR---IDT
Mflv_3397|M.gilvum_PYR-GCK FS-TDIKRTAVQYPRQGGKQDFGSFTSLPDVADEIVKMVSPHADGGVNGP
Mvan_3130|M.vanbaalenii_PYR-1 FA-TDIKRVAVQYPGQRGKQDFASFTSLPALADEVCKMVSPDKEGGVHGP
:. . ::** : . :: :* :
MSMEG_4514|M.smegmatis_MC2_155 PLCLFGHCMGAVVAFEFARVAERNGVAVEALWVSASEAPSAVAASPALPM
TH_4495|M.thermoresistible__bu PLDLFGHCMGAVVAFEFARVAERHAVPVGTLWASAGQAPSTVGEGPPLPT
MAV_2010|M.avium_104 PLTLFGHSMGGVVAFEFARVAEANGTPVRKLWVSAGPPPCVVGDMPELPT
MAB_2033|M.abscessus_ATCC_1997 QDVLFGHSLGGHIAFEVA-----QRLPVRKLVVSGIVAPSKVVDLGYRLL
Mb2953|M.bovis_AF2122/97 PVAFFGHSMGGMLAFEVALRYQSAGHRVLAFFVSACSAPGHIRYKQLQDL
Rv2928|M.tuberculosis_H37Rv PVAFFGHSMGGMLAFEVALRYQSAGHRVLAFFVSACSAPGHIRYKQLQDL
MLBr_02359|M.leprae_Br4923 AVAFFGHSMGGMLAFEVALRFQSAGYRLIALFVSACSAPGYIRYKQIKDF
MMAR_1778|M.marinum_M PVALFGHSMGGMLAFEVALRFEAAGYRVLALFLSACSAPGHIKYKQLKGY
MUL_2025|M.ulcerans_Agy99 PVALFGHSMGGMLAFEVALRFEAAGYRVLALFLSACSAPGHIKYKQLKGY
Mflv_3397|M.gilvum_PYR-GCK PVFFFGHSMGGLLAFEVARRFEDAGRPISALFVSAVAAPGLVGYDDIPD-
Mvan_3130|M.vanbaalenii_PYR-1 PIAFFGHSMGGLLAFEVARRFEEAGRPIAALFVSAVAAPGRVGYEDIPD-
:***.:*. :***.* : : *. .* :
MSMEG_4514|M.smegmatis_MC2_155 GESEIIAEMVDLGGTDPQLLADDDFVELLLTAVRADYEAFNRYSCDQGVT
TH_4495|M.thermoresistible__bu TDADVLADMVDLGGTDPRLLDDEDFVELLVMAVRADYRALSGYRCGPDER
MAV_2010|M.avium_104 THDGLLADIADLGGTDPELLADEEFSELLTTAVRADYQAFNGYDPSPDVR
MAB_2033|M.abscessus_ATCC_1997 NEAEGIERIAALGGLPPEILDDPSALSALFPVLRSDFTAGETYTPPPDAT
Mb2953|M.bovis_AF2122/97 SDREMLDLFTRMTGMNPDFFTDDEFFVGALPTLR-AVRAIAGYSCPPETK
Rv2928|M.tuberculosis_H37Rv SDREMLDLFTRMTGMNPDFFTDDEFFVGALPTLR-AVRAIAGYSCPPETK
MLBr_02359|M.leprae_Br4923 SDNDMLDLVVRMTGMNPDFFEDEEFRVGVLPTLR-AARIIAGYNCPPETT
MMAR_1778|M.marinum_M SDNEMLDLVARATGTDPEFFNDEEFRVGVLPTLR-AVRAIAGYSCPPENK
MUL_2025|M.ulcerans_Agy99 SDNEMLDLVARATGTDPEFFNDEEFRVGVLPTLR-AVRAIAGYSCPPENK
Mflv_3397|M.gilvum_PYR-GCK TDEGLLAAVTTITGADPEFMKNPEFAAAILPTLR-GLKAIANYTCPPDVT
Mvan_3130|M.vanbaalenii_PYR-1 DDEGLLAAVSSMTGANPEFMKNPEFAAAILPTLR-GLKAIANYTCPPDVT
. : . * * :: : . .:* *
MSMEG_4514|M.smegmatis_MC2_155 IAADIHALGGEQDHRISEDMLRGWQIHTEGAFTLSMFDGG-HFYINDYTD
TH_4495|M.thermoresistible__bu IRADIHALRGDRDHRISREWSDRWASHTSGRFTLSTFDGG-HFYLNDHLT
MAV_2010|M.avium_104 IGADIHVLGGHHDHRIATGVLRQWERHTAGSFAMSLYDGG-HFYLYDHVE
MAB_2033|M.abscessus_ATCC_1997 VACDITALIGDKDVLAPLERVSSWREHTRANFDLQVFRGGGHFYLEDHID
Mb2953|M.bovis_AF2122/97 LSCPIYAFIGDKDWIATQDDMDPWRDRTTEEFSIRVFPGD-HFYLNDNLP
Rv2928|M.tuberculosis_H37Rv LSCPIYAFIGDKDWIATQDDMDPWRDRTTEEFSIRVFPGD-HFYLNDNLP
MLBr_02359|M.leprae_Br4923 VSCPIYTYIGDKDWIATQEDMKPWRERTTGAFAIRVFPGD-HFYLNGNLS
MMAR_1778|M.marinum_M LSCPIYTFIGSKDWIATREDMEPWRERTTGDFSLREFPGD-HFYLNKNLP
MUL_2025|M.ulcerans_Agy99 LSCPIYTFIGSKDWIATREDMEPWRERTTGDFSLREFPGD-HFYLNKNLP
Mflv_3397|M.gilvum_PYR-GCK LSCPIHAYYGDDDEIATADKVMPWAGRTSAGFTARDFKGH-HFYLNDHLD
Mvan_3130|M.vanbaalenii_PYR-1 LSCPIHAFYGDDDEIATADKVKPWAERTTADFTAREFSGH-HFYLNDHLG
: . * . * * . * :* * : * ***:
MSMEG_4514|M.smegmatis_MC2_155 DVAELVNELW-------
TH_4495|M.thermoresistible__bu DVAALVSGR--------
MAV_2010|M.avium_104 TVAAQVNAG--------
MAB_2033|M.abscessus_ATCC_1997 EVSACLIQVCNVSPII-
Mb2953|M.bovis_AF2122/97 ELVSDIEDKTLQWHDRA
Rv2928|M.tuberculosis_H37Rv ELVSDIEDKTLQWHDRA
MLBr_02359|M.leprae_Br4923 ELVCDIEDKTLEWCDRA
MMAR_1778|M.marinum_M ELVSDIEIGTLQQFDQI
MUL_2025|M.ulcerans_Agy99 ELVSDIEIGTLQQFDQI
Mflv_3397|M.gilvum_PYR-GCK DVVPDIEQKVWSSLRE-
Mvan_3130|M.vanbaalenii_PYR-1 ELVPDLEQKLWSRLRE-
: :