For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSRIASQLGQPEHLTHALIILLTVAGVPSVYAGDEFGFRGLKEERYGVDDAVLPEFFSPPMQLDVFGVHA WALHQYLIGLRRRHP
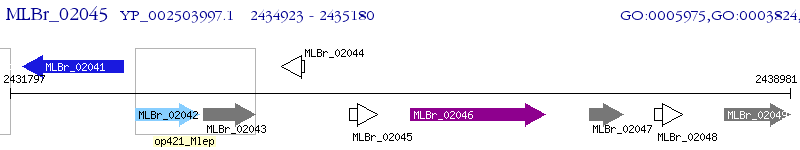
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. leprae Br4923 | MLBr_02045 | - | - | 100% (85) | hypothetical protein MLBr_02045 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3333 | - | 4e-24 | 60.00% (85) | alpha amylase, catalytic region |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_2753 | treS_1 | 2e-31 | 68.24% (85) | trehalose synthase TreS_1 |
| M. avium 104 | MAV_2842 | - | 4e-34 | 75.29% (85) | alpha-amylase 3 |
| M. smegmatis MC2 155 | MSMEG_3576 | - | 3e-24 | 63.53% (85) | alpha-amylase 3 |
| M. thermoresistible (build 8) | TH_2297 | - | 4e-22 | 53.93% (89) | alpha-amylase 3 |
| M. ulcerans Agy99 | MUL_3001 | treS_1 | 6e-30 | 65.88% (85) | trehalose synthase TreS_1 |
| M. vanbaalenii PYR-1 | Mvan_3059 | - | 2e-24 | 60.00% (85) | alpha amylase, catalytic region |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_3576|M.smegmatis_MC2_155 MTMPDWVTHAIWWQIYPLGFVGAHP-ADP---PPGPEEHRLRRIVEWLDH
TH_2297|M.thermoresistible__bu VTGPDWVRHTIWWQIYPLGFVGAHPWTDPTTGPPGPDEHRLRRVLDWLDH
Mflv_3333|M.gilvum_PYR-GCK MSDPAWVEHVLWWQVYPLGFVGVHP-ADT---APEPGEHRLRRIVDWLDY
Mvan_3059|M.vanbaalenii_PYR-1 MTEPDWVDHVVWWQVYPLGFVGAYP-ADP---PPPPGEHRLRRIVDWLDY
MLBr_02045|M.leprae_Br4923 --------------------------------------------------
MAV_2842|M.avium_104 MTGPDWVQHVIWWQVYPLGFVGAYPLPQSSE-PPQPEHHRLRRLVDWFDH
MMAR_2753|M.marinum_M ------MQHAIWWQVYPLGFVGAFP---ANQ-PPDPGEHRLRRLVGWFDH
MUL_3001|M.ulcerans_Agy99 MSKPTWVQHAIWWQVYPLGFVGAFP---ANQ-PPNPGEHRLRRLVGWFDH
MSMEG_3576|M.smegmatis_MC2_155 AVELGASGLALGPVFASRTHGYDTTDHLRIDPRLGDDADFDHVVAEAHRR
TH_2297|M.thermoresistible__bu VVDLGASGLALGPVFASRSHGYDTTDHYRIDPRLGTDADFDELVEQAHRR
Mflv_3333|M.gilvum_PYR-GCK AVELGASGVALGPVFHSRTHGYDTIDHYRVDPRLGDDSDFDHLIEESHSR
Mvan_3059|M.vanbaalenii_PYR-1 AIELGASGLALGPVFASRTHGYDTTDHYRIDPRLGEDGDFDHLVAEARDR
MLBr_02045|M.leprae_Br4923 --------------------------------------------------
MAV_2842|M.avium_104 AIELGASGIALGPIFTSRTHGYDTTDHYRIDPRLGDDADFDHLIAQAHRR
MMAR_2753|M.marinum_M AVELGASGVALGPIFDSRTHGYDTTDHFRIDPRLGDEGDFDHLVAEAHRR
MUL_3001|M.ulcerans_Agy99 AVELGASGVALGPIFDSRTHGYDTTDHFRIDPRLGDEGDFDHLVAEAHRR
MSMEG_3576|M.smegmatis_MC2_155 GLRVLLDGVFNHVGTDFARYRQALGGDADSAA-WFRHRDGKFEVFEGHGE
TH_2297|M.thermoresistible__bu GLRVLLDGVFHHVGTDFARYRDAAAG--GSSG-WFRRRGDRFETFEGHDE
Mflv_3333|M.gilvum_PYR-GCK GLRVLLDGVFNHVGTEFPRYREAVDG--GDRS-WFATRGDDFATFEGHEG
Mvan_3059|M.vanbaalenii_PYR-1 GLRILLDGVFNHVGTDFPRYRDAAAG--GDSA-WFRRRGGDFATFEGHGD
MLBr_02045|M.leprae_Br4923 --------------------------------------------------
MAV_2842|M.avium_104 GLRVLLDGVFNHVGVDFPRYRAAIDSDDHAAARWFRGSRGRFHTFEGHDG
MMAR_2753|M.marinum_M GLRVLLDGVFNHVGMDFHRYRHAAD--DPSSAGWFRGRAGNFRTFEGHSE
MUL_3001|M.ulcerans_Agy99 GLRVLLDGVFNHVGVDFHSYRHAAD--DPSSAGWFRGRAGNFRTFEGHSG
MSMEG_3576|M.smegmatis_MC2_155 LIALNHDNPEVVNYTAGVMRHWLGRGADGWRLDAAYAVPDRFWAQVLPIV
TH_2297|M.thermoresistible__bu LIALDHDNPEVAAYTVDVLRHWLNRGADGWRLDAAYAVPDRFWARVLPDV
Mflv_3333|M.gilvum_PYR-GCK LITLDHDNADVVDYTVDVMSHWLARGADGWRLDAAYAVPDRFWAQVLPRV
Mvan_3059|M.vanbaalenii_PYR-1 LIALDHRSAEVVDYTVDVMSHWLGRGADGWRLDAAYAVPDSFWAGVLPRV
MLBr_02045|M.leprae_Br4923 --------------------------------------------------
MAV_2842|M.avium_104 LITLNHDNPEVVDYTVDVMAHWLGRGADGWRLDAAYAVPETFWAATLPRV
MMAR_2753|M.marinum_M LITLNHANPEVADYVVDVMAHWLQRGADGWRLDAAYAVPQQFWAKVLPRV
MUL_3001|M.ulcerans_Agy99 LITLNHANPEVADYVVDVMAHWLQRGADGWRLDAAYAVPQQFWAKVLPRV
MSMEG_3576|M.smegmatis_MC2_155 RSDFGDAWFVGEVIHGDYAATVAASTFDSVTQYELWKAVWSALNDGNFHE
TH_2297|M.thermoresistible__bu RRSHPEAWFVGEVIHGDYAATVSAASFDSVTQYELWKAVWSSLNDGNFFE
Mflv_3333|M.gilvum_PYR-GCK RQAHPAAWFVGEVIHGDYVARVRDSGFDSVTQYELWKAVWSSINDANFHE
Mvan_3059|M.vanbaalenii_PYR-1 RERHPQAWFVGEVIHGDYPARVRDSGFDSVTQYELWKAIWSSLNDGNFHE
MLBr_02045|M.leprae_Br4923 --------------------------------------------------
MAV_2842|M.avium_104 RDRYPQAWFVGELIHGDYAAVVRAAGFDSATQYELWKAIWSSLNDGNFYE
MMAR_2753|M.marinum_M RDRYPRAWFVGEIIHGDYAAAVQGAGFDSVTQYELWKAIWSGLNDGNFFE
MUL_3001|M.ulcerans_Agy99 RDRYPQAWFVGEIIHGDYAAAVQGAGFDSVTQYELWKAIWSGLNDGNFFE
MSMEG_3576|M.smegmatis_MC2_155 LDWALQRHNDFLASFVPMTFVGNHDVTRIASQLSDARHLEHALVILFTTG
TH_2297|M.thermoresistible__bu LDWALQRHNDFLAVFTPLTFIGNHDVTRIASRLADRRHVEHAVVLLLTVG
Mflv_3333|M.gilvum_PYR-GCK LDWALTRHNEFLDTFVPLTFIGNHDVTRIASRLTDPAHVEHALVVLLTTG
Mvan_3059|M.vanbaalenii_PYR-1 LDWALTRHDEFLDTFVPMTFVGNHDVTRIASQLTDPTHVEHALVLLMTVG
MLBr_02045|M.leprae_Br4923 -------------------------MSRIASQLGQPEHLTHALIILLTVA
MAV_2842|M.avium_104 LDWALQRHNAFLGRFSPLTFIGNHDVTRIASQLNNPAHLAHALVLLLTVG
MMAR_2753|M.marinum_M LDWALQRHNEFVASFAPLTFIGNHDVTRIASRLERPEHLAHALVILMTVG
MUL_3001|M.ulcerans_Agy99 LDWALQRHNEFVASFAPLTFIGNHDVTRIASRLERPEHLAHALVILMTVG
::****:* *: **:::*:*..
MSMEG_3576|M.smegmatis_MC2_155 GIPSVYAGDEWGHQGVKEERRGGDDAVRPEFGERP-----GPDAPGADVL
TH_2297|M.thermoresistible__bu GVPSIYAGDEAGYLGVKEERFSGDDAIRPEFGSPPIVSGAGENTVAADLV
Mflv_3333|M.gilvum_PYR-GCK GTPSVYAGDEVAYRGVKEERFGGDDAVRPEFRSPF----DGVGEQGHQMF
Mvan_3059|M.vanbaalenii_PYR-1 GTPSVYAGDEVAYRGVKEERFGGDDAVRPEFGSPF----EGVGEHGHQVF
MLBr_02045|M.leprae_Br4923 GVPSVYAGDEFGFRGLKEERYGVDDAVLPEFFSPP----MQLDVFGVHAW
MAV_2842|M.avium_104 GVPSVYAGDELGFRGVKEERFGGDDAVRPEFGSPP----IPLDDFGTEVW
MMAR_2753|M.marinum_M GIPSIYAGDEFGWRAVKEERYGGDDAIRPEFGSPP----LQLTHDQASVW
MUL_3001|M.ulcerans_Agy99 GIPSIYAGDEFGWRAVKEERCGGDDAIRPEFGSPP----FELTHDQASVW
* **:***** . .:**** . ***: *** .
MSMEG_3576|M.smegmatis_MC2_155 RLHQYLIGLRRRHPWLHSATTTPLLLTNTQYVYRSAGAAGALVVALNLDD
TH_2297|M.thermoresistible__bu RLHQHLIGLRRRNPWLHTATTAPLHLTNTQYVYESRSGEHALVVALNIDT
Mflv_3333|M.gilvum_PYR-GCK RSHQHLIGLRRRHPWLHEARTTALSVTNTGYVYITRSGDDALVVALNVAD
Mvan_3059|M.vanbaalenii_PYR-1 RSHQHLIGLRRRNPWLHTARTTPLRVTNRGYVYTTRHGDDALVVALNIDD
MLBr_02045|M.leprae_Br4923 ALHQYLIGLRRRHP------------------------------------
MAV_2842|M.avium_104 RLHQYLIGLRRRHPWLHAATTTALLLQNRHYVYETRSGDEALLIVLNIGD
MMAR_2753|M.marinum_M ALHQYLVGLRRRYPWLHTASTSALRLRNRQYVYQTRSADNALVVILNADD
MUL_3001|M.ulcerans_Agy99 ALHQYLVGLRRRYPWLHTASTSALRLRNRQYVYQTRSADNALVVILNADD
**:*:***** *
MSMEG_3576|M.smegmatis_MC2_155 APLRVALPELGLASGVVVAGSKAPPQEQIEVTEVPPHGWLVIEPR-----
TH_2297|M.thermoresistible__bu APLPLRLSEIGLGAGRVVAGSGAPPAADLTNVDVAPHGWLIIEPAR----
Mflv_3333|M.gilvum_PYR-GCK EPLPVALSGLGLGHAEIVAGSGAPPAEVVPDIVVPPHGWVILNPR-----
Mvan_3059|M.vanbaalenii_PYR-1 DAMPVSLAEFGLARAEVVAGSGAPPAQVVADTVVPAHGWLILAPR-----
MLBr_02045|M.leprae_Br4923 --------------------------------------------------
MAV_2842|M.avium_104 EPLRVSLDELGGRRGRVIGGSAAPPPEVLDTVVVEPQGWRILSPI-----
MMAR_2753|M.marinum_M EPLRMSLPDLGIPRAEVLAGSAAPPQEVTDEVLVEGHGWRILQPA-----
MUL_3001|M.ulcerans_Agy99 EPLRMSLPDLGIPHAEVLAGSAAPPQEVTDEVLVEGHGWRILQPASRFSC
MSMEG_3576|M.smegmatis_MC2_155 ----------
TH_2297|M.thermoresistible__bu ----------
Mflv_3333|M.gilvum_PYR-GCK ----------
Mvan_3059|M.vanbaalenii_PYR-1 ----------
MLBr_02045|M.leprae_Br4923 ----------
MAV_2842|M.avium_104 ----------
MMAR_2753|M.marinum_M ----------
MUL_3001|M.ulcerans_Agy99 ASVSGSSRRA