For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ATLAESPEPKSGASRAGVLGEEAGYHKGLKPRQLQMIGIGGAIGTGLFLGAGGRLAKAGPGLFLVYAVCG VFVFLILRALGELVLHRPSSGSFVSYAREFFGEKAAYVVGWLYFLDWAMTAIVDTTAIATYLHRWTIFTA LPQWTLALLALAVVLVMNLISVEWFGELEFWAALIKVCALMAFLVVGTIFLGGRYPVDGHNTGLSLWTSH GGLFPTGVAPLIVVSSGVMFAYAAVELVGTAAGETVEPKKIMPRAINSVIARIAIFYVGSVILLALLLPY SAFKASESPFVTFFSKVGFYGAGDLMNIVVLTAALSSLNAGLYATGRVMHSIAINGSGPKFTARMSKXGV PYGGILLAAVICLCGVALNAFNPGQAFEIVLSVAALGIIAGWGTIVLCQLRLHKMAKAGIMRRPRFRMPL APYSGYLTLAFLFAVLVVMAFDKPIGTWTVASLIVIVPALIAGWYSIRKRVMTIARERMGYTGPFPAIAN PPVKSSD*
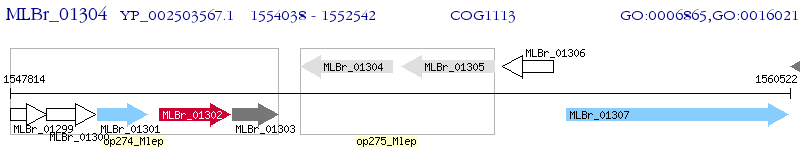
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. leprae Br4923 | MLBr_01304 | ansP1 | - | 100% (498) | putative L-asparagine permease |
| M. leprae Br4923 | MLBr_01305 | ansP2 | 0.0 | 99.20% (498) | putative L-asparagine permease |
| M. leprae Br4923 | MLBr_01974 | - | 1e-12 | 22.22% (396) | putative permease |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2151 | ansP1 | 0.0 | 82.75% (487) | L-asparagine permease ansP1 |
| M. gilvum PYR-GCK | Mflv_1998 | - | 7e-74 | 36.47% (447) | amino acid permease-associated region |
| M. tuberculosis H37Rv | Rv2127 | ansP1 | 0.0 | 83.16% (487) | L-asparagine permease ansP1 |
| M. abscessus ATCC 19977 | MAB_2914 | - | 0.0 | 65.12% (473) | L-asparagine permease 1 (L-asparagine transport) |
| M. marinum M | MMAR_4815 | ansP1 | 0.0 | 71.19% (486) | L-asparagine permease AnsP1 |
| M. avium 104 | MAV_4626 | - | 8e-77 | 38.00% (450) | gaba permease |
| M. smegmatis MC2 155 | MSMEG_6196 | - | 3e-78 | 36.32% (468) | gaba permease |
| M. thermoresistible (build 8) | TH_0471 | - | 6e-18 | 24.03% (437) | POSSIBLE CATIONIC AMINO ACID TRANSPORT INTEGRAL MEMBRANE |
| M. ulcerans Agy99 | MUL_0383 | ansP1 | 0.0 | 70.16% (486) | L-asparagine permease AnsP1 |
| M. vanbaalenii PYR-1 | Mvan_1740 | - | 2e-18 | 23.17% (423) | amino acid permease-associated region |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1998|M.gilvum_PYR-GCK ---------------------MSAAPTLKKALNQ----RQLTMIAIGGVI
MAV_4626|M.avium_104 ---------------------MAGTSQLRQGLSQ----RQLSMIALGGVI
MSMEG_6196|M.smegmatis_MC2_155 --------------MLESRSDLSSSDEPRSGLARSLKTRHMTMIAIGGAI
Mb2151|M.bovis_AF2122/97 ---------------MSAASQRVDAFGEEAGYHKGLKPRQLQMIGIGGAI
Rv2127|M.tuberculosis_H37Rv ---------------MSAASQRVGAFGEEAGYHKGLKPRQLQMIGIGGAI
MLBr_01304|M.leprae_Br4923 ------MATLAESPEPKSGASRAGVLGEEAGYHKGLKPRQLQMIGIGGAI
MMAR_4815|M.marinum_M ----------MRTPSSPTHPDDIALASEDAGYHKSLKPRQLQMIAIGGAI
MUL_0383|M.ulcerans_Agy99 ----------MRTPSSPTHPDDIALASEDAGYHKSLKPRQLQMIAIGGAI
MAB_2914|M.abscessus_ATCC_1997 -------------MTSPATYDTSAPAPEEQGYHKGLNNRQVQMIGIGGAI
TH_0471|M.thermoresistible__bu --------MTDRLRVKSVEQSIADTDEPDSRLRKDLSWWDLTVFGVSVVI
Mvan_1740|M.vanbaalenii_PYR-1 MVQEPGAQTVGRRRTKSVEQSIADTDEPSTRLRKELNWWDLTVFGVSVVI
: .: ::.:. .*
Mflv_1998|M.gilvum_PYR-GCK GAGLFVGSG-VVIADTGPGAFLTYGMAGVLIIMVMRMLAEMAVANPSTGS
MAV_4626|M.avium_104 GAGLFVGSG-VVIKDTGPAAFLTYALCGLLIVLVMRMLGEMAAANPSTGS
MSMEG_6196|M.smegmatis_MC2_155 GAGLFVGSG-AVIKTAGPAAIVSYVVAGALVLCTLRMLGEMVVAKPRTGA
Mb2151|M.bovis_AF2122/97 GTGLFLGAS-GRLAKAGPGLFLVYGVCGVFVFLILRALGELVLHRPSSGS
Rv2127|M.tuberculosis_H37Rv GTGLFLGAG-GRLAKAGPGLFLVYGVCGVFVFLILRALGELVLHRPSSGS
MLBr_01304|M.leprae_Br4923 GTGLFLGAG-GRLAKAGPGLFLVYAVCGVFVFLILRALGELVLHRPSSGS
MMAR_4815|M.marinum_M GTGLFLGAG-GRLASAGPALFLVYAVCGVFAFLILRALGELVLHRPSSGS
MUL_0383|M.ulcerans_Agy99 GTGLFLGAG-GRLASAGPALFLVYAVCGVFAFLILRALSELVLHRPSSGS
MAB_2914|M.abscessus_ATCC_1997 GTGLFMGAG-GRLHSAGPGLFLVYAVCGVFVFFILRALGELILHRPSSGS
TH_0471|M.thermoresistible__bu GAGIFTVTASTAGNITGPAISVSFIIAAIACGLAALCYAEFASTVPVAGS
Mvan_1740|M.vanbaalenii_PYR-1 GAGIFTITASTAANITGPAISLSFIIAAIACGLAALCYAEFASTVPVAGS
*:*:* :. :**. : : :.. .*: * :*:
Mflv_1998|M.gilvum_PYR-GCK FADYSRNALGHWAGFSVGWLYWYFWVIVVGFEAIAGAKIVQYW---MDVP
MAV_4626|M.avium_104 FADYAAAALGRWAGFSVAWLYWYFWVIVVGFEAVAGGKVLNYW---FHAP
MSMEG_6196|M.smegmatis_MC2_155 FADYARIGIGPWAGFTLGWLYWYYYVIIIAVEAVAGAQILAVW---VGLP
Mb2151|M.bovis_AF2122/97 FVSYAREFFGEKAAYAVGWMYFLHWAMTSIVDTTAIATYLQRWTIFTVVP
Rv2127|M.tuberculosis_H37Rv FVSYAREFFGEKAAYAVGWMYFLHWAMTSIVDTTAIATYLQRWTIFTVVP
MLBr_01304|M.leprae_Br4923 FVSYAREFFGEKAAYVVGWLYFLDWAMTAIVDTTAIATYLHRWTIFTALP
MMAR_4815|M.marinum_M FVSYAREFFGEKAAYVAGWMYFLNWAMTSIVDSTAIATYFHFWRVFEGIP
MUL_0383|M.ulcerans_Agy99 FVSYAREFFGEKAAYVAGWMYFLNWAMTSIVDSTAIATYFHFWRVFEGIP
MAB_2914|M.abscessus_ATCC_1997 FVSYAREFLGEKAAYVAGWMYFFNWAATAIVDVTAIALYMHYWSAFKAIP
TH_0471|M.thermoresistible__bu AYTFSYATFGEFVAWIIGWDLILEFSVAAAVVAKGWSSYLGEVFEFAGAT
Mvan_1740|M.vanbaalenii_PYR-1 AYTFSYATFGEFVAWIIGWDLILEFAVAAAVVAKGWSSYLGTVFGFAGGT
:: :* ..: .* : . . . . .
Mflv_1998|M.gilvum_PYR-GCK LWLS-------ALIFMVLMTATNLFSVASYGEFEFWFAGIKVAAIIAFIA
MAV_4626|M.avium_104 LWVL-------SLGLMLLMTATNLFSVSSFGEFEFWFAGIKVATIVIFLV
MSMEG_6196|M.smegmatis_MC2_155 LWLM-------AMLLMAALTLTNLISVRWFGEFEFWFSGIKVAAIVSFIV
Mb2151|M.bovis_AF2122/97 QWIL-------ALIALTVVLSMNLISVEWFGELEFWAALIKVLALMAFLV
Rv2127|M.tuberculosis_H37Rv QWIL-------ALIALTVVLSMNLISVEWFGELEFWAALIKVLALMAFLV
MLBr_01304|M.leprae_Br4923 QWTL-------ALLALAVVLVMNLISVEWFGELEFWAALIKVCALMAFLV
MMAR_4815|M.marinum_M QWAL-------ALFALGIALSMNLISVTLFGEIEFWAALIKVVSLLTFLV
MUL_0383|M.ulcerans_Agy99 QWAL-------ALFALGIALSMNLISETLFGEIEFWAALIKVVSLLTFRV
MAB_2914|M.abscessus_ATCC_1997 QWSI-------ALIALVIVLTMNTISVKLFGEMEFWAALIKVVAIMGFLV
TH_0471|M.thermoresistible__bu TRLGPITLDWGALLIIGFVTVLLATGTKLSANVSLAITVIKVGVVLLVVI
Mvan_1740|M.vanbaalenii_PYR-1 TEVAGFQLDWGALVIIALVTLILVMGTKLSAGVSLAITAVKVSVVLLVVI
:: : . . ..: : :** :: .
Mflv_1998|M.gilvum_PYR-GCK VGAAFVLG---------LWPGKGADFSNLTSHGGFMP------LGFGAIT
MAV_4626|M.avium_104 VGAAYVIG---------LVPGHHDGLTNLVSHGGFFP------RGVGAVF
MSMEG_6196|M.smegmatis_MC2_155 VGLLWVFG---------VWPGDTNGLTNLVAHGGFAP------NGFGSIF
Mb2151|M.bovis_AF2122/97 VGTVFLAGR-------YPVDGHSTGLSLWNNHGGLFP------TSWLPLL
Rv2127|M.tuberculosis_H37Rv VGTVFLAGR-------YPVDGHSTGLSLWNNHGGLFP------TSWLPLL
MLBr_01304|M.leprae_Br4923 VGTIFLGGR-------YPVDGHNTGLSLWTSHGGLFP------TGVAPLI
MMAR_4815|M.marinum_M VGTIFLAGR-------YPVDGHSTGLGLLAHGGGLFP------YGLLPLV
MUL_0383|M.ulcerans_Agy99 VGTIFLAGR-------YPVDGHSTGLGLLAHGGGLFP------YGLLPLV
MAB_2914|M.abscessus_ATCC_1997 IGTVFLAGR-------FTVEGASTGPSVIADNGGLLP------TGLMALV
TH_0471|M.thermoresistible__bu VGAFFIKAANYTPFIPPAEAGGEAGRSLDQSLFSLITGAEGSSYGWYGLL
Mvan_1740|M.vanbaalenii_PYR-1 VGAFYIKAENFSPFVPAPEEGGSGGTGTEQSLFSLLTGAEGSHYGWYGVL
:* :: . * . .: . . :
Mflv_1998|M.gilvum_PYR-GCK VGIVTVIFSMVGAEIATIAAAESQDPERAVAKAANSVILRILIFYVGAVL
MAV_4626|M.avium_104 AAIVVVIFSMVGAEIVTVAAAESRDPRRAVQKATRSVVTRIGIFFVGSVF
MSMEG_6196|M.smegmatis_MC2_155 GAVVVVIFAFGGAEIVTIAAAESAEPAKSVARATTGVIWRIILFYVGSIF
Mb2151|M.bovis_AF2122/97 IVTSGVVFAYSAVELVGTAAGETAEPEKIMPRAINSVVARIAIFYVGSVA
Rv2127|M.tuberculosis_H37Rv IVTSGVVFAYSAVELVGTAAGETAEPEKIMPRAINSVVARIAIFYVGSVA
MLBr_01304|M.leprae_Br4923 VVSSGVMFAYAAVELVGTAAGETVEPKKIMPRAINSVIARIAIFYVGSVI
MMAR_4815|M.marinum_M VVTSGVVFAYAAVELVGTAAGETAEPEKVMPRAINSVIARIALFYVGSLF
MUL_0383|M.ulcerans_Agy99 VVTSGVVFAYAAVELVGTAAGETAEPEKVMPRAINSVIARIALFYVGSLF
MAB_2914|M.abscessus_ATCC_1997 IVTSGVVFAYSAVELVGIAAGETENPDKVMPRAINSVVFRVAVFYVGSLI
TH_0471|M.thermoresistible__bu AGASIVFFAFIGFDIIATTAEETKNPQRDVARGIMATLGIVTVLYVAVSL
Mvan_1740|M.vanbaalenii_PYR-1 AGASIVFFAFIGFDIVATTAEETRNPQRDVPRGILASLAIVTVLYVAVAV
*.*: . :: :* *: :* : : :. . : : :::*.
Mflv_1998|M.gilvum_PYR-GCK LLVLIVPWNDESVS--ASPFVAAFNAMGIPYADHVMNAVVLTAVLSCLNS
MAV_4626|M.avium_104 LLVAILPWDSVELG--ASPYVAALKHLGLAGADQVMNAVVLTAVLSCLNS
MSMEG_6196|M.smegmatis_MC2_155 LVVCILPWNDASVL--SSPYVAALEKLQIPGAGLIMNIVILTAVLSVLNS
Mb2151|M.bovis_AF2122/97 LLALLLPYTAYKAG--ESPFVTFFSKIGFHGAGDLMNIVVLTAALSSLNA
Rv2127|M.tuberculosis_H37Rv LLALLLPYTAYKAG--ESPFVTFFSKIGFHGAGDLMNIVVLTAALSSLNA
MLBr_01304|M.leprae_Br4923 LLALLLPYSAFKAS--ESPFVTFFSKVGFYGAGDLMNIVVLTAALSSLNA
MMAR_4815|M.marinum_M LLGLLLPYTDYKAG--ESPFVTFFDKIGVTGAGSIMNVVVMTAAFSSLNA
MUL_0383|M.ulcerans_Agy99 LLGLLLPYTDYKAG--ESPFVTFFDKIGVTGAGSIMNVVVMTAAFSSLNA
MAB_2914|M.abscessus_ATCC_1997 LLALLLPYGTYKAG--ESPFVTFFSRIGVPYAGDVMNFVVLTAALSSLNA
TH_0471|M.thermoresistible__bu VLTGMVHYTELRAAGENANLATAFAANGIDWAAKVISIGALAGLTTVVIV
Mvan_1740|M.vanbaalenii_PYR-1 VLTGMVSYTELRDA-ETQNLATAFSLNGVDWAAKVISIGALAGLTTVVIV
:: :: : .: : . * ::. ::. : :
Mflv_1998|M.gilvum_PYR-GCK GMYTASRMLFVLAARREAPAALVKVTNRGVPANAILASSVVGFLCVIAAA
MAV_4626|M.avium_104 GLYTASRMLFVLAERGEAPARLVRLSGRGVPHIAILCSSAVGFLCVVMAR
MSMEG_6196|M.smegmatis_MC2_155 SIYTSSRMLRVLASHGDAPQWLVRTNTRGVPTRAILASTVVGWVSVVLAY
Mb2151|M.bovis_AF2122/97 GLYSTGRVMHSIAMSGSAPRFTARMSKSGVPYGGIVLTAVITLFGVALNA
Rv2127|M.tuberculosis_H37Rv GLYSTGRVMHSIAMSGSAPRFTARMSKSGVPYGGIVLTAVITLFGVALNA
MLBr_01304|M.leprae_Br4923 GLYATGRVMHSIAINGSGPKFTARMSKXGVPYGGILLAAVICLCGVALNA
MMAR_4815|M.marinum_M GLYSTGRILRSMAMNGSAPSFTARMSQRGVPYGGICLTAAIGLIGVLLNG
MUL_0383|M.ulcerans_Agy99 GLYSTGRILRSMAMNGSAPSFTARMSQRGVPYGGICLTAAIGLIGVLLNG
MAB_2914|M.abscessus_ATCC_1997 GLYSTGRILRSLAMNGSAPKVLSRMTSGGVPFMGIAVTGALTLVGIFMNL
TH_0471|M.thermoresistible__bu LVLGQTRVLFAMSRDGLLPRPLSKTGPRGTPVR---ITVLVGALVAIAAS
Mvan_1740|M.vanbaalenii_PYR-1 LVLGQTRVLFAMSRDGLLPRQLAKTGEHGTPVR---ITLIVGAVVAVTAT
: *:: :: * : *.* : :
Mflv_1998|M.gilvum_PYR-GCK VSPDTIFAFLLNSSGAIILFVYLLIAISQIVLR---YRTPDSKLRVKMWL
MAV_4626|M.avium_104 VAPNTVFLFLLNSSGAIILFVYWLIALSQIVLR---RRTPAERLSVKMWF
MSMEG_6196|M.smegmatis_MC2_155 IAPDSLFLFLVNSCGVVAIFMYVMIGVSELRVRRAIEREDPAKLTLKMWF
Mb2151|M.bovis_AF2122/97 FKPGEAFEIVLNMSALGIIAGWATIVLCQLRLHKLANAGIMQRPRFRMPF
Rv2127|M.tuberculosis_H37Rv FKPGEAFEIVLNMSALGIIAGWATIVLCQLRLHKLANAGIMQRPRFRMPF
MLBr_01304|M.leprae_Br4923 FNPGQAFEIVLSVAALGIIAGWGTIVLCQLRLHKMAKAGIMRRPRFRMPL
MMAR_4815|M.marinum_M VVPEMAFEIVLNIAALGILASWATIVCCQIQLFRWSKQGIVYRPSFRMWG
MUL_0383|M.ulcerans_Agy99 VVPEMAFEIVLNIAALGILASWATIVCCQIQLFRWSKQGIVYRPSFRMWG
MAB_2914|M.abscessus_ATCC_1997 VIPAEAFETALDLAALGIISSWATIVICQIQLYRWSQRGILQRGSFRMPG
TH_0471|M.thermoresistible__bu VFPVGNLEEMVN---IGTLFAFALVSAGVIVLR---RTRPDLKRGFRVPF
Mvan_1740|M.vanbaalenii_PYR-1 VFPIGKLEEMVN---IGTLFAFVLVSAGVLVLR---RTRPDLKRGFRTPW
. * : :. : : : : : : .:
Mflv_1998|M.gilvum_PYR-GCK FPVLSGLTALGILAILVQMFIDTALRSQLVLSLLSWAVVIALFAANKWFV
MAV_4626|M.avium_104 FPVLSVLTLAGITAVLVQMAFDATARSQLWLSLLSWAVVVALY-----FV
MSMEG_6196|M.smegmatis_MC2_155 FPYLPIAVIACFVLVLVAMLFDADQRVQLLASVVSLAVVLLAYALRKRHG
Mb2151|M.bovis_AF2122/97 SPYSGYLTLLFLLVVLVTMASDKPIGTWTVATLIIVIPALTAGWYLVRKR
Rv2127|M.tuberculosis_H37Rv SPYSGYLTLLFLLVVLVTMASDKPIGTWTVATLIIVIPALTAGWYLVRKR
MLBr_01304|M.leprae_Br4923 APYSGYLTLAFLFAVLVVMAFDKPIGTWTVASLIVIVPALIAGWYSIRKR
MMAR_4815|M.marinum_M APYTGYLTLVFLFAVLVLMGFSYPIGTWTVATLGLIIPMLIGGWFLVRDR
MUL_0383|M.ulcerans_Agy99 APYTGYLTLVFLFAVLVLMGFSYPIDTWTVATLGLIIPMLIGGWFLVRDR
MAB_2914|M.abscessus_ATCC_1997 APYTGYLTLAFLAAVVVLMCYEN---AWNLIAIAVLVPVLTAGWYLCRGR
TH_0471|M.thermoresistible__bu VPWLPLAAIAACLWLMLNLTGLTWVRFLGWMALGLVLYFFYGRHHSMLGR
Mvan_1740|M.vanbaalenii_PYR-1 VPLLPILAIAACVWLMLNLTGLTWIRFLIWMAIGVVVYFLYGRRHSLVGR
* . ::: : :: .
Mflv_1998|M.gilvum_PYR-GCK KKRPAEGDSGAPTGPPHRVLVLANRTVDSDELLAELSRIGADQTAEYLVV
MAV_4626|M.avium_104 AR--SRGRVAAR--------------------------------------
MSMEG_6196|M.smegmatis_MC2_155 QVPATDTKQAARTDE-----------------------------------
Mb2151|M.bovis_AF2122/97 VMAVARERLGHTGPFPAVANPPVRSRD-----------------------
Rv2127|M.tuberculosis_H37Rv VMAVARERLGHTGPFPAVANPPVRSRD-----------------------
MLBr_01304|M.leprae_Br4923 VMTIARERMGYTGPFPAIANPPVKSSD-----------------------
MMAR_4815|M.marinum_M VLEIARRRIGYTGEFPVVANPGPMDKK-----------------------
MUL_0383|M.ulcerans_Agy99 VLEIARRRIGYIGEFPVVANPGPMDKK-----------------------
MAB_2914|M.abscessus_ATCC_1997 VLQLARERIGYTGAYPVIAHTPMLDDPPGESHR-----------------
TH_0471|M.thermoresistible__bu GMTDISARRTG---------------------------------------
Mvan_1740|M.vanbaalenii_PYR-1 RMG-----------------------------------------------
Mflv_1998|M.gilvum_PYR-GCK VPASAVDTGAAATHGPRDVTEATQEAATARLESTLAALRGKNLTVQGELG
MAV_4626|M.avium_104 --------------------------------------------------
MSMEG_6196|M.smegmatis_MC2_155 --------------------------------------------------
Mb2151|M.bovis_AF2122/97 --------------------------------------------------
Rv2127|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_01304|M.leprae_Br4923 --------------------------------------------------
MMAR_4815|M.marinum_M --------------------------------------------------
MUL_0383|M.ulcerans_Agy99 --------------------------------------------------
MAB_2914|M.abscessus_ATCC_1997 --------------------------------------------------
TH_0471|M.thermoresistible__bu --------------------------------------------------
Mvan_1740|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_1998|M.gilvum_PYR-GCK DYRPLRALDHAVAEFGPDQIVIATLPPEFSMWHRFDVVDRARAQFTVPVT
MAV_4626|M.avium_104 --------------------------------------------------
MSMEG_6196|M.smegmatis_MC2_155 --------------------------------------------------
Mb2151|M.bovis_AF2122/97 --------------------------------------------------
Rv2127|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_01304|M.leprae_Br4923 --------------------------------------------------
MMAR_4815|M.marinum_M --------------------------------------------------
MUL_0383|M.ulcerans_Agy99 --------------------------------------------------
MAB_2914|M.abscessus_ATCC_1997 --------------------------------------------------
TH_0471|M.thermoresistible__bu --------------------------------------------------
Mvan_1740|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_1998|M.gilvum_PYR-GCK HVVATSAVGNQVPR
MAV_4626|M.avium_104 --------------
MSMEG_6196|M.smegmatis_MC2_155 --------------
Mb2151|M.bovis_AF2122/97 --------------
Rv2127|M.tuberculosis_H37Rv --------------
MLBr_01304|M.leprae_Br4923 --------------
MMAR_4815|M.marinum_M --------------
MUL_0383|M.ulcerans_Agy99 --------------
MAB_2914|M.abscessus_ATCC_1997 --------------
TH_0471|M.thermoresistible__bu --------------
Mvan_1740|M.vanbaalenii_PYR-1 --------------