For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LRVVDLSSAPQDGPASPPGVIVAVGSADQIARSEFWLDAATFTLTEDACTDRRAVTVDSVSTSLDELTAR CGRWPHASAICDDVLRAVDPAGPAARGLLTESLAYSTLQAGPEFARWLDERGPARMPEIADPVLARRDGD VLRIAFNRPQRHNAFSTDARAALLEALAVAQLDPSVTGIVLSGNGPSFCSGGDLAEFGTFADPASAHLAR TRHSPALVLDALTARFGRACRAEVHGRVLGSGLEMAAFCGWVSARDDAVFGLPELALGLIPGAGGTVSIT RRIGRWRTAYLVLSGRTVGAATALSWGLVDEVEAATASS*
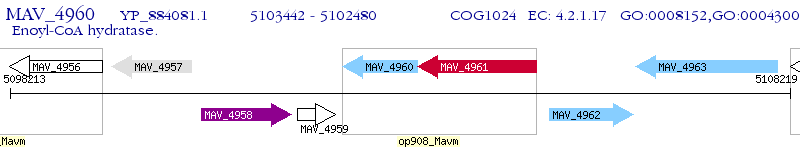
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_4960 | - | - | 100% (320) | enoyl-CoA hydratase/isomerase family protein |
| M. avium 104 | MAV_1195 | - | 1e-22 | 35.83% (187) | enoyl-CoA hydratase |
| M. avium 104 | MAV_2601 | - | 1e-21 | 34.43% (183) | enoyl-CoA hydratase/isomerase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1099c | echA8 | 3e-23 | 35.83% (187) | enoyl-CoA hydratase |
| M. gilvum PYR-GCK | Mflv_0448 | - | 1e-120 | 74.49% (294) | enoyl-CoA hydratase/isomerase |
| M. tuberculosis H37Rv | Rv1070c | echA8 | 3e-23 | 35.83% (187) | enoyl-CoA hydratase |
| M. leprae Br4923 | MLBr_02402 | - | 1e-21 | 35.63% (174) | enoyl-CoA hydratase |
| M. abscessus ATCC 19977 | MAB_3382c | - | 6e-21 | 33.51% (188) | enoyl-CoA hydratase/isomerase |
| M. marinum M | MMAR_0453 | echA8_4 | 1e-139 | 77.29% (317) | enoyl-CoA hydratase, EchA8_4 |
| M. smegmatis MC2 155 | MSMEG_0259 | - | 1e-115 | 72.66% (278) | enoyl-CoA hydratase/isomerase |
| M. thermoresistible (build 8) | TH_4433 | - | 1e-118 | 70.95% (296) | PUTATIVE enoyl-CoA hydratase/isomerase |
| M. ulcerans Agy99 | MUL_1103 | echA8_4 | 1e-133 | 75.08% (317) | enoyl-CoA hydratase, EchA8_4 |
| M. vanbaalenii PYR-1 | Mvan_0219 | - | 1e-120 | 73.47% (294) | enoyl-CoA hydratase/isomerase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0453|M.marinum_M MFGVVDLSRAPDATRAAGEISAPGVTVAHGWASELAEAEPWLSAATFTLA
MUL_1103|M.ulcerans_Agy99 MFGVVDLSRAPDATRAAGEISAPGVTVAHGWASELAEAEPWLSAATFTLA
MAV_4960|M.avium_104 MLRVVDLSSAPQD----GPASPPGVIVAVGSADQIARSEFWLDAATFTLT
Mflv_0448|M.gilvum_PYR-GCK MNTLVELA--------------GGIVVGVG---TPSDA------ATFTLT
Mvan_0219|M.vanbaalenii_PYR-1 MNSPVELG--------------GGVRVAVG---TPSDD------ATFTVT
MSMEG_0259|M.smegmatis_MC2_155 ---MIELP--------------GGIRIGLIDDPDDAED------ATFTLS
TH_4433|M.thermoresistible__bu -VSILQLA--------------DGVVIGLG---EPVDD------ATFTLT
Mb1099c|M.bovis_AF2122/97 ----MTYET---------------ILVE------------------RDQR
Rv1070c|M.tuberculosis_H37Rv ----MTYET---------------ILVE------------------RDQR
MLBr_02402|M.leprae_Br4923 ----MKYDT---------------ILVD------------------GYQR
MAB_3382c|M.abscessus_ATCC_199 ---MIEPEF---------------VSVRT---------------APEHPG
: : :
MMAR_0453|M.marinum_M EGPCADRRVITVESVSQTLAELRQRCQRWPQASAVCDDVLRTIDPDGPTL
MUL_1103|M.ulcerans_Agy99 EGPCADRRVITVESVSQTLAELRQRVPRWPQARAVCDDVLRTIDPDGSTL
MAV_4960|M.avium_104 EDACTDRRAVTVDSVSTSLDELTARCGRWPHASAICDDVLRAVDPAGPAA
Mflv_0448|M.gilvum_PYR-GCK DEDTDDRRAVRVASVDEALAELTERCLRWPQAAAVCDDVLRAFDPQGCTF
Mvan_0219|M.vanbaalenii_PYR-1 EDDVDDRRAVTVTSVPDTLAELEERCRRWPQAAAVCDDVLRAFDPGGSTF
MSMEG_0259|M.smegmatis_MC2_155 EAATDDRRVVTVASIADTLELLRQRCERRPQTAAVCDDVLRSADPDASTF
TH_4433|M.thermoresistible__bu EKPTDDRRAVTVESVQDTLDLLRERCGKWPQTAAVCDDVLRSIDPDGPTR
Mb1099c|M.bovis_AF2122/97 VGIITLNRPQALNALNSQVMNEVTSAATELDDDPDIGAIIITGSAKAFAA
Rv1070c|M.tuberculosis_H37Rv VGIITLNRPQALNALNSQVMNEVTSAATELDDDPDIGAIIITGSAKAFAA
MLBr_02402|M.leprae_Br4923 VGIITLNRPQALNALNSQMMNEITNAAKELDIDPDVGAILITGSPKVFAA
MAB_3382c|M.abscessus_ATCC_199 IGTIALSRP-PTNTFTRQMYRELAQAAAEVNARADIRVVIVYGGHEIFCA
* :. : . . :: .
MMAR_0453|M.marinum_M AGVLTESLAYSTLQAGPEFARWLDERGPADLPDIADPVQTERDDDTLRIR
MUL_1103|M.ulcerans_Agy99 AGVLTESLAYSTLQAGPEFARWLDERGPADLPDIADPVQTERDDDTLRIR
MAV_4960|M.avium_104 RGLLTESLAYSTLQAGPEFARWLDERGPARMPEIADPVLARRDGDVLRIA
Mflv_0448|M.gilvum_PYR-GCK TGVITESLAYSTLQAGAEFARWLDARGPARVPEIADPVVAERVDDVLTIR
Mvan_0219|M.vanbaalenii_PYR-1 AGVITESLAYSTLQSGSEFARWLAERGPARVPDISDPVVAERHDGRLTIR
MSMEG_0259|M.smegmatis_MC2_155 SGVITESLAYSTLQSGAEFARWLDERGPATVPQLPDPVRAHRDGNTLHVR
TH_4433|M.thermoresistible__bu SGVITESLAYSTLQSGSEFARWLAERGPKTVPQLPDPVLATRDGDTLRVV
Mb1099c|M.bovis_AF2122/97 GADIKEMADLTFADAFT--ADFFATWG--KLAAVRTPTIAAVAGYALGGG
Rv1070c|M.tuberculosis_H37Rv GADIKEMADLTFADAFT--ADFFATWG--KLAAVRTPTIAAVAGYALGGG
MLBr_02402|M.leprae_Br4923 GADIKEMASLTFTDAFD--ADFFSAWG--KLAAVRTPMIAAVAGYALGGG
MAB_3382c|M.abscessus_ATCC_199 GDDLAELSELSADEMARSAGDRQAALT--AVAELAKPTIAAITGYALGSG
: * : : . :. : * : . *
MMAR_0453|M.marinum_M FNRPQRHNAFSGDARAALLEALTVAQLDPSVSGIVLSGNGPSFCSGGDLA
MUL_1103|M.ulcerans_Agy99 FNRPQRHIAFSGDARAALLEALTVAQLDPSVSGIVLSGNGPSFCSGGDLA
MAV_4960|M.avium_104 FNRPQRHNAFSTDARAALLEALAVAQLDPSVTGIVLSGNGPSFCSGGDLA
Mflv_0448|M.gilvum_PYR-GCK FNRPQRHNAFSTDARAALLDALEVARLDPSVTGMVLTGNGPSFCSGGDLA
Mvan_0219|M.vanbaalenii_PYR-1 FNRPQRHNAFSTDARAALLEALEVARLDPSVDEVVLAGNGPSFCSGGDLA
MSMEG_0259|M.smegmatis_MC2_155 FNRPQRHNAFSTDARAALLEALEVARLDPSVDEVVLAGNGQSFCSGGDLA
TH_4433|M.thermoresistible__bu FNRPQRHNAFSTDARAALLEALEVARLDPSVTEVVLTGNGPSFGSGGDLA
Mb1099c|M.bovis_AF2122/97 CELAMMCDVLIAADTAKFGQPEIKLGVLPGMGGSQRLTRAIGKAKAMDLI
Rv1070c|M.tuberculosis_H37Rv CELAMMCDVLIAADTAKFGQPEIKLGVLPGMGGSQRLTRAIGKAKAMDLI
MLBr_02402|M.leprae_Br4923 CELAMMCDLLIAADTAKFGQPEIKLGVLPGMGGSQRLTRAIGKAKAMDLI
MAB_3382c|M.abscessus_ATCC_199 LELALTADWRIAGDNVKVGSPEILVGMIPGGGGTVRLARVVGLSRAKEMV
: . . . .. : *. . . . ::
MMAR_0453|M.marinum_M EFGTFADPASAHLARTRHSPALALAALTARLGRACRAEVHGRVLGSGLEM
MUL_1103|M.ulcerans_Agy99 ELGTFADPASAHLARTRHSPALALAALTARLGRACRAEVHGRVLGSGLEM
MAV_4960|M.avium_104 EFGTFADPASAHLARTRHSPALVLDALTARFGRACRAEVHGRVLGSGLEM
Mflv_0448|M.gilvum_PYR-GCK EFGTFTDPASAHLARTRYSPALVLAEITARLGDKCRAEIHGQVQGSGLEM
Mvan_0219|M.vanbaalenii_PYR-1 EFGTFTDPAGAHLARTRHSPALVLDEITARLGRGCRAELHGQVQGSGLEM
MSMEG_0259|M.smegmatis_MC2_155 EFGSFADPAAAHLARTRYSPALVLDELTARLGRRCRAHVHGQVLGSGLEM
TH_4433|M.thermoresistible__bu EFGSFDDPAGAHLARTRHSPALVLAELTERLGPRCRAEVHGLVLGSGLEM
Mb1099c|M.bovis_AF2122/97 LTGRTMDAAEAERSGLVSRVVPADDLLTEARATATTISQMSASAARMAKE
Rv1070c|M.tuberculosis_H37Rv LTGRTMDAAEAERSGLVSRVVPADDLLTEARATATTISQMSASAARMAKE
MLBr_02402|M.leprae_Br4923 LTGRTIDAAEAERSGLVSRVVLADDLLPEAKAVATTISQMSRSATRMAKE
MAB_3382c|M.abscessus_ATCC_199 FSGRFVGADEARGAGLIDQVVAPDDVYTEAASWARRFVDGPPLALAAAKE
* .. *. : . . . :
MMAR_0453|M.marinum_M AAFCGWVQAHPDSVFGLPELSLGLIPGAGGTVSVTRRIGRWRTAYLVLSG
MUL_1103|M.ulcerans_Agy99 AAFCGWVQAQPDSVFGLPELSLGLIPGAGGAVSVTRRIGRWRTAYLALSG
MAV_4960|M.avium_104 AAFCGWVSARDDAVFGLPELALGLIPGAGGTVSITRRIGRWRTAYLVLSG
Mflv_0448|M.gilvum_PYR-GCK AAFCGTVSCDAGAVLGLPELALGLIPGAGGTVSITRRIGRWRTAYLVLSG
Mvan_0219|M.vanbaalenii_PYR-1 AAFCGWVSCDADAVLGLPELTLGLIPGAGGTVSVTRRIGRWRTAYLVLSG
MSMEG_0259|M.smegmatis_MC2_155 ACFCGWVSCDPDAKLGLPELELGLVPGAGGTVSITRRIGRWRTAYLVLSG
TH_4433|M.thermoresistible__bu ASFCGHVTAHPDAVFGLPELELGLIPGAGGCVSITRRIGRWRTAYLVLTG
Mb1099c|M.bovis_AF2122/97 AVNRAFESSLSEGLLYERRLFHSAFATEDQSEGMAAFIEKRAPQFTHR--
Rv1070c|M.tuberculosis_H37Rv AVNRAFESSLSEGLLYERRLFHSAFATEDQSEGMAAFIEKRAPQFTHR--
MLBr_02402|M.leprae_Br4923 AVNRSFESTLAEGLLHERRLFHSTFVTDDQSEGMAAFIEKRAPQFTHR--
MAB_3382c|M.abscessus_ATCC_199 AIDHGFDGPLAAGLDKERELFCQVFATQDRADGVTAQLEIGPGTARFRGA
* . . .* . . .:: :
MMAR_0453|M.marinum_M RTIDAGTARDWGLVDSIAVTGD--
MUL_1103|M.ulcerans_Agy99 RTIDAGTARDWGLVDSIAVTGD--
MAV_4960|M.avium_104 RTVGAATALSWGLVDEVEAATASS
Mflv_0448|M.gilvum_PYR-GCK RSIDAATALRWGLVDAVG------
Mvan_0219|M.vanbaalenii_PYR-1 RPIDAGTALRWGLVDAVG------
MSMEG_0259|M.smegmatis_MC2_155 RTIDPTTALSWGLVDEIVSVAAPA
TH_4433|M.thermoresistible__bu RRIDAVTALRWGLVDAVSS-----
Mb1099c|M.bovis_AF2122/97 ------------------------
Rv1070c|M.tuberculosis_H37Rv ------------------------
MLBr_02402|M.leprae_Br4923 ------------------------
MAB_3382c|M.abscessus_ATCC_199 ------------------------