For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MLVTMSERHAPEPVGNPLRGIFVVNRVAGRWPFALRAAICMAVPVLVGWSAGNTSAGLIATIGGFTSLYG SGRPYLNRGVFLGGVAVTFAVVVALGDWASRHVWLGVATVTVIAMVTVLVCHALSVGPPGAYMFVLACAA GTGAPGTADLGPVRMALLVLAGGSFAWCVHMLGALIRPRGPERAAVRDAALAIAEYIDSIGGDANAEGQA RHRAATLLHTAWVTLVTFQPVQPKPDQALYRLRVVNRRLHVLLADAMRAADAGTAPPPGGAAFARELAAI PPLTDEHGAEGVPLGRPGAGQLLRQALRPGSMSLRLAARVAVAVAISGAIAVGFAEALAIDRAYWAMAAA VLMLHQGFAWVRTVTRSVERMVGTWLGLGVAGAILAVHPQGVWLALTIGVLQFVIEMYVVRNYTLAVVFI TPAALTISSGGAAITDLGALLWARGVDTLIGCAVALLAYRATQRMRAPTALSTAITRTLDAVDATLPHLG AGAATTGAARTHRRDLHVRAMALLPSYDAAVGGSAAQRRAAERLWPTVVAAEQLAYRTLAACWALERDGD VDAARDTADALAEQASDVRGRLDEGQ
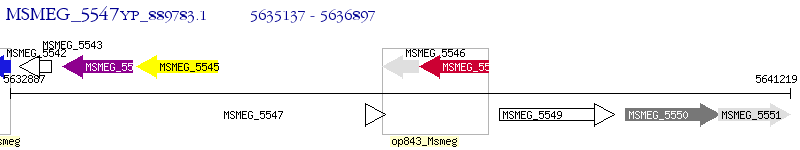
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5547 | - | - | 100% (586) | hypothetical protein MSMEG_5547 |
| M. smegmatis MC2 155 | MSMEG_3414 | - | 3e-11 | 31.88% (207) | hypothetical protein MSMEG_3414 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3327 | - | 2e-06 | 25.13% (597) | protein of unknown function DUF893, YccS/YhfK |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2268c | - | 6e-44 | 28.50% (565) | hypothetical protein MAB_2268c |
| M. marinum M | MMAR_0945 | - | 7e-09 | 25.44% (283) | hypothetical protein MMAR_0945 |
| M. avium 104 | MAV_4566 | - | 2e-11 | 25.18% (413) | hypothetical protein MAV_4566 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0945|M.marinum_M MSRRDDVQGRPAVDTRIPESMAARLQRAQSWAIGSDPGLLRLRMATRTTA
MAV_4566|M.avium_104 -------------------------------MIGSDPGLLRLRMATRTTA
MSMEG_5547|M.smegmatis_MC2_155 --------------------------------------MLVTMSERHAPE
MAB_2268c|M.abscessus_ATCC_199 --------------------------------------MTPPAPESPDPL
Mflv_3327|M.gilvum_PYR-GCK ---------------------------------MIMGLTVRSVLPDAAAA
.
MMAR_0945|M.marinum_M ALGCSLLVLYLLTMATGQPLTVALIGVVITMIAARSVDDPDSRQQQITMA
MAV_4566|M.avium_104 ALGLSLLALWLLTRATGQPLTVALLGVVITMVASRSVNEPDPRQQRITMA
MSMEG_5547|M.smegmatis_MC2_155 PVGNPLRGIFVVNRVAGR--WPFALRAAICMAVPVLVGWSAGNTSAGLIA
MAB_2268c|M.abscessus_ATCC_199 PQIARARSILFSAPPAGRR-WSIGLRAAAAVAIPGTLVVAAGYPNAALYV
Mflv_3327|M.gilvum_PYR-GCK LRSLAAVGVIAVIAALWGPPGSATSAVAVAAVAGATAFQDSPRSRIPLVY
: ..
MMAR_0945|M.marinum_M LLPVPAALSISVAALLAPHLIAAEVAFVVIVFAAVYVRRFGPRGRALG--
MAV_4566|M.avium_104 LLPVPAAVAITAAAVLAPHPVAGDVVFVLIVFAAAYIRRFGARGRALG--
MSMEG_5547|M.smegmatis_MC2_155 TIGGFTSLYGSGRPYLNRGVFLGGVAVTFAVVVALGDWASRHVWLGVA--
MAB_2268c|M.abscessus_ATCC_199 TYGAFAVLYGEGRPYRVRAAVVATAGIALMLVTAVGVVVGSCAPGGVTNK
Mflv_3327|M.gilvum_PYR-GCK LVSGLCGVAVLLGTAVSGLPVLFLLLCALWCAGVAMLWAHGPNTGLIG--
: . . .. . :
MMAR_0945|M.marinum_M -----MVAFMAYFFSLYLRAGVAQLPWMIGAVLVGTLCTFVMSTYVLPDR
MAV_4566|M.avium_104 -----MVAFMSYFFTLYLRARVSELPWMIGAVAVGTVCTYVMTSHVLPDR
MSMEG_5547|M.smegmatis_MC2_155 -----TVTVIAMVTVLVCHALSVGPPGAYMFVLACAAGTGAPGTADLG--
MAB_2268c|M.abscessus_ATCC_199 AATILTLAAIAIPAVYTVDALRLGPPGALFFVLVCGGALGATEWGVAP--
Mflv_3327|M.gilvum_PYR-GCK ----AASGALMVAATSAGTPVSVGAALGTAALAVVGGLSQAVVLAVWP--
: . . : . .
MMAR_0945|M.marinum_M PESVLRATIRSLRARMAIVVDTAADTVRAGRLDERRRRRMRARIARLNET
MAV_4566|M.avium_104 PERVLRATIRALRARMAIVIDTTAEAVRTGRLDERRRRRMRARTVRLNET
MSMEG_5547|M.smegmatis_MC2_155 ------------PVRMALLVLAGGSFAWCVHMLGALIRPRGPERAAVRDA
MAB_2268c|M.abscessus_ATCC_199 -------------VRILVCASLGAGAAVLVSMTGAVIDRQKPERVATHRA
Mflv_3327|M.gilvum_PYR-GCK ------------PQRWRAQRDAVVAAYRSLAADARRLADGRDDGWAVDTE
*
MMAR_0945|M.marinum_M ALLVQSQIEDKTHPATLWPGISAEQLAPWLFDAELSVEWVAIAGHRAMAI
MAV_4566|M.avium_104 ALMVQSQIEDRADPGTLWPGVTAEQLAPWLLDAELAIEWVATAGRRAAAL
MSMEG_5547|M.smegmatis_MC2_155 ALAIAEYIDSIGG------DANAEGQARHRAATLLHTAWVTLVTFQPVQP
MAB_2268c|M.abscessus_ATCC_199 VEAVDVYAR----------PGNATVDARHAAGAAISSAWTALHDAGLSSR
Mflv_3327|M.gilvum_PYR-GCK PLVALREAFTDVS------GPTLRRPAVYRGWYALPERIAATLTDMANGP
. * : .:
MMAR_0945|M.marinum_M AEEIPAATRIELADTLTQLAWAIRGPRTEGLHQAASRAQRLLDGQPAPAP
MAV_4566|M.avium_104 GAELPEAVRAELVSALTQLGRAIRLPEPGGLRDAASRARRLLD----AAT
MSMEG_5547|M.smegmatis_MC2_155 KPDQ----------ALYRLRVVNR-----RLHVLLADAMRAAD-----AG
MAB_2268c|M.abscessus_ATCC_199 SDSP--------------------------LVTTMLDTHRRFT-------
Mflv_3327|M.gilvum_PYR-GCK RSEALSRVLEEAADTMAAVADTSRAG-RVGTHVSIGRFDSVVTEIDGPES
.
MMAR_0945|M.marinum_M TDDPSQGAVRRLGLAIVAAATATSEVRELIGDAT-AAPPDPDNND----V
MAV_4566|M.avium_104 DDRPAGTEVRRLALAIINAATATADVRAIVDGAAGAAVPDVAGSERPPAA
MSMEG_5547|M.smegmatis_MC2_155 TAPPPGGAAFARELAAIPPLTDEHGAEGVPLGRP----------------
MAB_2268c|M.abscessus_ATCC_199 ----------EISVGNKMVLDHLIPGDHVLVVRP----------------
Mflv_3327|M.gilvum_PYR-GCK GTVRRLSALLHEVAAMRLGDFVPSTPDAVRLRRP---------------E
. : .
MMAR_0945|M.marinum_M VQPVPEDTAADGGPGEAGLAPTTRQAIQVSIAASLAIVTGELVSPLHWYW
MAV_4566|M.avium_104 AGVAAEEPAEQEEEQPAGLRPTTRQAIQVSVAASLAIITGELVSPARWYW
MSMEG_5547|M.smegmatis_MC2_155 --GAGQLLRQALRPGSMSLRLAARVAVAVAISGAIAVGFAEALAIDRAYW
MAB_2268c|M.abscessus_ATCC_199 --SIWYRLRRSVRFRSHATITAGRAGIACLGAG----VLSAAVGLTRPQW
Mflv_3327|M.gilvum_PYR-GCK LPTSVRSAMDTVRGHLNQDSPVLRHAVRVGAAVGLGGGIAQYAGIPYGYW
. * .: : . . *
MMAR_0945|M.marinum_M AVIAAFVVFAGTNSWGETLTKGWQRLLGTVLGVPCGVLVATVVAGDKVFS
MAV_4566|M.avium_104 AVIAAFVIFAGTNSWGETLTKGWQRLLGTMLGVPAGVLVATLLTGHEAAA
MSMEG_5547|M.smegmatis_MC2_155 AMAAAVLMLHQGFAWVRTVTRSVERMVGTWLGLGVAGAILAVHPQGVWLA
MAB_2268c|M.abscessus_ATCC_199 AILSALVIVHMGPDRLHGTVRGLHRFAGTVVGLALFAVLYQLSLTGYALI
Mflv_3327|M.gilvum_PYR-GCK IPLTALLVLRP--ETAHTYTRCVGRVAGTAAGVVVASAVLVPFAPAPVVS
:*.::. . .: *. ** *: :
MMAR_0945|M.marinum_M VVMIFACLFFAVYFMQVTYSLMTFWITTMLALLYGLLGKFTFGVLMLRIE
MAV_4566|M.avium_104 LAGIFVCLFCAFYFMTVTYSLMTFWITTMLALLYGLLGQFSFGVLMLRIE
MSMEG_5547|M.smegmatis_MC2_155 LTIGVLQFVIEMYVVRNYTLAVVFITPAALTISSGGAAITDLGALLW---
MAB_2268c|M.abscessus_ATCC_199 AAIATLQFCTELFLPRNYAVAVTFVTPIALLSAG---VVTLHGSVTSIVR
Mflv_3327|M.gilvum_PYR-GCK LGFAVVAVGIAYLVSGVGYIAMSGAVAAAAVFLLDVARAGTPSSLTDPLI
. . : . . :
MMAR_0945|M.marinum_M ETAIGAVIGVAVAILVLPTTIGTVVRRD---TRAFLTTLSELIEISIATM
MAV_4566|M.avium_104 ETALGAVIGATVAVVVLPTNTRTAIRAD---TRAFLTSLSALIEACAAAM
MSMEG_5547|M.smegmatis_MC2_155 ARGVDTLIGCAVALLAYRATQRMRAPTA---LSTAITRTLDAVDATLPHL
MAB_2268c|M.abscessus_ATCC_199 DRLAETLIGVAVAMAS----IYLVAPHA---HRRTFAFTEARIRRAALGL
Mflv_3327|M.gilvum_PYR-GCK ATIAGGALAVIAHVALPDDELTRLAQRAGELLKTEIDYAATVIKGYVHER
:. . : : :
MMAR_0945|M.marinum_M FDDDVTDSPTERARELDRDLQQFRVTAKP-MLAGVAGFAGRRSIQHSVRL
MAV_4566|M.avium_104 SGG--AASPSEQARQLDRDLQQFRVTAKP-LLAGVAGLAGRRGIRRALRI
MSMEG_5547|M.smegmatis_MC2_155 GAG---AATTGAARTHRRDLHVRAMALLPSYDAAVGGSAAQR--------
MAB_2268c|M.abscessus_ATCC_199 VAAARIQAAHDAAYAHARDLSFELEGAIR---AGVDSAHDEP--------
Mflv_3327|M.gilvum_PYR-GCK ENATEEMTAAWQRVSRARSAFEATTGAMRLESRDFRLWLRSYRTAIN---
:. *. .
MMAR_0945|M.marinum_M LTICDRYARILARRSEQYLD-PGCPHGLSDAITTAAAQIRRNIDALVTAL
MAV_4566|M.avium_104 FTACDRYGRALARSSEQYRGSPGPGPQLAQAFSAAAAQTRRNLDALLEAI
MSMEG_5547|M.smegmatis_MC2_155 ----------------------RAAERLWPTVVAAEQLAYRTLAACWALE
MAB_2268c|M.abscessus_ATCC_199 ----------------------EWTQVHWPAHAALIHHGYDLVAACWATP
Mflv_3327|M.gilvum_PYR-GCK ----------------SLTSSCTTLEAHMPTRPAGVDREFAITVDEFVEA
: :
MMAR_0945|M.marinum_M DGTQDVAITP--AADSLDAAEAVVRQHADALDRADPGSNPRHLLAAVNSL
MAV_4566|M.avium_104 DSGHPPTLVS--AADELDAAETAARQHDSDGD-GETRPDVRRFLTAVHAL
MSMEG_5547|M.smegmatis_MC2_155 RDGDVDAARD--TADALAEQASDVRGRLDEGQ------------------
MAB_2268c|M.abscessus_ATCC_199 ------------PGQMLADPDRWERRFRGEA-------------------
Mflv_3327|M.gilvum_PYR-GCK LCGDPPTAASPWTVDAAELEAAAQRLREAAPRERAEDGSSRVLVAEATAI
. : *
MMAR_0945|M.marinum_M RRIEGAVVNAAVHFGADADLMAHDQPG-
MAV_4566|M.avium_104 RQIERAVISTATNLGGREDVRTATAAPR
MSMEG_5547|M.smegmatis_MC2_155 ----------------------------
MAB_2268c|M.abscessus_ATCC_199 ----------------------------
Mflv_3327|M.gilvum_PYR-GCK TRSLTAVAISPVPISAR-----------