For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAKLKPYYEESQATYDISDDFFALFLDPNMVYTCAYFENDDMTLEQAQLAKLDLGLDKLNLEPGMTLLDV GCGWGGAVVRALEKYDVNVIGITLSRNHYARTKERLAAIPTTRRAEARLQGWEEFDEPVDRIISFEAFDA FKKERWPAFWDWAYKSLPNDSRMLMHSIFTYPQSQWKERGIPITMSDLRFFHFLGKEIFPGGQMCGEADI VDLSQASGFSLEQVQYLRPHYARTLDTWAANLEANREQAIAIQSQEVYDRFMRYLTGCAELFRKGISNVA QYTLTK
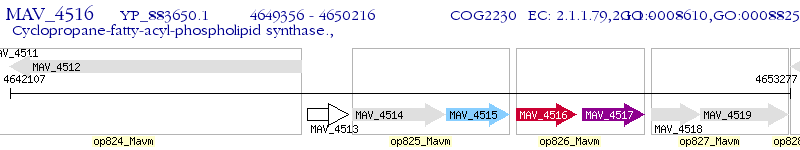
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_4516 | - | - | 100% (286) | methoxy mycolic acid synthase 1 |
| M. avium 104 | MAV_4680 | - | e-116 | 68.53% (286) | methoxy mycolic acid synthase 1 |
| M. avium 104 | MAV_0130 | - | 1e-98 | 57.95% (283) | cyclopropane-fatty-acyl-phospholipid synthase 1 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0664c | mmaA1 | 1e-124 | 73.43% (286) | methoxy mycolic acid synthase |
| M. gilvum PYR-GCK | Mflv_1800 | - | 5e-95 | 56.18% (283) | cyclopropane-fatty-acyl-phospholipid synthase |
| M. tuberculosis H37Rv | Rv0645c | mmaA1 | 1e-124 | 73.43% (286) | methoxy mycolic acid synthase |
| M. leprae Br4923 | MLBr_01900 | mmaA1 | 1e-123 | 71.68% (286) | methyl mycolic acid synthase 1 |
| M. abscessus ATCC 19977 | MAB_4088c | - | 1e-104 | 62.28% (289) | mycolic acid synthase UmaA1 |
| M. marinum M | MMAR_0980 | mmaA1 | 1e-118 | 69.93% (286) | methoxy mycolic acid synthase 1 Mma1 |
| M. smegmatis MC2 155 | MSMEG_0913 | - | 1e-106 | 62.46% (285) | methoxy mycolic acid synthase 1 |
| M. thermoresistible (build 8) | TH_3763 | - | 1e-116 | 67.37% (285) | PUTATIVE Cyclopropane-fatty-acyl-phospholipid synthase |
| M. ulcerans Agy99 | MUL_4538 | umaA | 1e-113 | 64.69% (286) | mycolic acid synthase UmaA |
| M. vanbaalenii PYR-1 | Mvan_0803 | - | 1e-107 | 63.96% (283) | cyclopropane-fatty-acyl-phospholipid synthase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0664c|M.bovis_AF2122/97 MA------------------KLRPYYEESQSAYDISDDFFALFLD-PTWV
Rv0645c|M.tuberculosis_H37Rv MA------------------KLRPYYEESQSAYDISDDFFALFLD-PTWV
MLBr_01900|M.leprae_Br4923 MA------------------KLRPYYEESQSAYDISDDFFALFLD-PTWV
MMAR_0980|M.marinum_M MA------------------ELKPYFEDSQATYDISDEFFGLFLD-PTWV
MAV_4516|M.avium_104 MA------------------KLKPYYEESQATYDISDDFFALFLD-PNMV
MSMEG_0913|M.smegmatis_MC2_155 MSN--------------VESELAPYYEESQSIYDISNEFFALFLG-PTMG
Mvan_0803|M.vanbaalenii_PYR-1 MSN--------------AVPDLSPYYEESQSIYDVSNEFFALFLG-PTMG
MAB_4088c|M.abscessus_ATCC_199 MS------------------DLKPYYEESQSIYDISDEFYGLFLDEETMG
MUL_4538|M.ulcerans_Agy99 MT------------------ELRPFYEESQSIYDVSDEFFALFLD-PTMA
TH_3763|M.thermoresistible__bu MTN--------------TSAELEPFFEESQSIYDVSDDFFALFLG-PTMG
Mflv_1800|M.gilvum_PYR-GCK MTGRTAEPPTGHDTTPPTAEDMKPHFEEVQAHYDLSDVFFGLFQD-PSRT
*: .: *.:*: *: **:*: *:.** . .
Mb0664c|M.bovis_AF2122/97 YTCAYFERDDMTLEEAQLAKVDLALDKLNLEPGMTLLDVGCGWGGALVRA
Rv0645c|M.tuberculosis_H37Rv YTCAYFERDDMTLEEAQLAKVDLALDKLNLEPGMTLLDVGCGWGGALVRA
MLBr_01900|M.leprae_Br4923 YTCAYFERDDMTLEEAQLAKLDLALDKLNLAPGMTLLDVGCGWGGALVRA
MMAR_0980|M.marinum_M YTCAYFERDDMTLEEAQLAKLDLALDKLHLEPGMTLLDVGCGWGGALVRA
MAV_4516|M.avium_104 YTCAYFENDDMTLEQAQLAKLDLGLDKLNLEPGMTLLDVGCGWGGAVVRA
MSMEG_0913|M.smegmatis_MC2_155 YTCGYYEREDMTLDESQNAKFDLALGKLDLKPGMTLLDVGCGWGGALERA
Mvan_0803|M.vanbaalenii_PYR-1 YTCSYFEREDMTLDEAQIAKFDLALGKLNLEPGMTLLDVGCGWGGALRRA
MAB_4088c|M.abscessus_ATCC_199 YTCAYFERDDLTLAEAQIAKFDLALGKLNLEPGMTVLDIGCGWGACLDRA
MUL_4538|M.ulcerans_Agy99 YTCAYFERDDMTLEEASNAKFDLALGKLNLEPGMTLLDIGCGWGGALQRA
TH_3763|M.thermoresistible__bu YTCAYFERDDMTLDEAQIAKFDLALGKLNLEPGMTLLDIGCGWGGALQLA
Mflv_1800|M.gilvum_PYR-GCK YSCAYYERDDMTLAEAQIAKIDLALGKLNLEPGMTLLDIGCGWGSVLQRA
*:*.*:*.:*:** ::. **.**.*.**.* ****:**:*****. : *
Mb0664c|M.bovis_AF2122/97 VEKYDVNVIGLTLSRNHYERSKDRLAAIG--TQRRAEARLQGWEEFEENV
Rv0645c|M.tuberculosis_H37Rv VEKYDVNVIGLTLSRNHYERSKDRLAAIG--TQRRAEARLQGWEEFEENV
MLBr_01900|M.leprae_Br4923 VEKYDVNVIGLTLSRNHCARSKARLAEIL--TKRHAEARLQGWEEFEEKV
MMAR_0980|M.marinum_M VEKYDVNVIGLTLSRNHCQRSQDRLAALG--TKRRAEARLQGWEEFDEQV
MAV_4516|M.avium_104 LEKYDVNVIGITLSRNHYARTKERLAAIP--TTRRAEARLQGWEEFDEPV
MSMEG_0913|M.smegmatis_MC2_155 VTKYDVNVIGITLSKAQSDYARERLAKI--ETNRSVEVRLQGWEEFNEPV
Mvan_0803|M.vanbaalenii_PYR-1 VEKYDVNVIGITLSRKQFEYSKALMAGL--DTERKAEVRLQGWEEFDEPV
MAB_4088c|M.abscessus_ATCC_199 MRKFDVNVIGITLSKNQSEYSRKRLAQVAAETGRSAEIRMQGWEEFNDKV
MUL_4538|M.ulcerans_Agy99 VEKYDVNVIGITLSRNQFDYSKAKMAKIP--TERTVEVRLQGWEEFEDKV
TH_3763|M.thermoresistible__bu LEKYDVNVIGITLSRNQYEYSKARLAQID--TARTAEVRLQGWEQFDEPV
Mflv_1800|M.gilvum_PYR-GCK VEKYDVNVVGLTLSRNQCKFAQDLLDGLD--TERSRRILLRGWEQFDEPV
: *:****:*:***: : :: : : * * . ::***:*:: *
Mb0664c|M.bovis_AF2122/97 DRIVSFEAFDAFKKERYLTFFERSYDILPDDGRMLLHSLFTYDRRWLHEQ
Rv0645c|M.tuberculosis_H37Rv DRIVSFEAFDAFKKERYLTFFERSYDILPDDGRMLLHSLFTYDRRWLHEQ
MLBr_01900|M.leprae_Br4923 DRIVSFEALDAFKKERYPAFFERSYNILPDDGRMLLHSLFTYDRRWLHEQ
MMAR_0980|M.marinum_M DRIVSFEAFDAFRKERYGAFFQRSYDILPAGGSLLLHSLFTYDRRWLHEQ
MAV_4516|M.avium_104 DRIISFEAFDAFKKERWPAFWDWAYKSLPNDSRMLMHSIFTYPQSQWKER
MSMEG_0913|M.smegmatis_MC2_155 DRIVSIGAFEAFKAERYPVFFERAYSILPDDGRMLLHTILAHTQKFFREN
Mvan_0803|M.vanbaalenii_PYR-1 DRIVSIGAFEAFKAERYPLFFERAYDILPADGRMLLHTILAHTQKFFRDN
MAB_4088c|M.abscessus_ATCC_199 DRIVTIGAFEAFKQERYPIFFERAYDILPNDGRMLLHTILAHTQQFFREN
MUL_4538|M.ulcerans_Agy99 DRIVTIGAFEAFKMERYAAFFERSYDILPDDGRMLLHTILTYTQKQFHEM
TH_3763|M.thermoresistible__bu DRIVSIGAFEAFKKPRYAAFFEMAHKALPADGRMLLHTILAHPQTYFREA
Mflv_1800|M.gilvum_PYR-GCK DRIISIEAIEAFPQERYAPFFKMCSSVLPEGGRFVLQAILGHPLKKWPEL
***::: *::** *: *:. . . ** .. ::::::: : :
Mb0664c|M.bovis_AF2122/97 GIALTMSDLRFLKFLRESIFPGGELPSEPDIVDNAQAAGFTIEHVQLLQQ
Rv0645c|M.tuberculosis_H37Rv GIALTMSDLRFLKFLRESIFPGGELPSEPDIVDNAQAAGFTIEHVQLLQQ
MLBr_01900|M.leprae_Br4923 GIPLTMGDLRFLKFLRESIFPGGELPSQPDIVDNAEAAGFSVEQIQLMQP
MMAR_0980|M.marinum_M GIALTMSDLRFLKFLRESIFPGGEVPSESDIVDNAQAAGFAVEQIQLMQQ
MAV_4516|M.avium_104 GIPITMSDLRFFHFLGKEIFPGGQMCGEADIVDLSQASGFSLEQVQYLRP
MSMEG_0913|M.smegmatis_MC2_155 GIKLTISDLKFMKFIGEEIFPGGQLPAVEDIEKLAADSGFKLERIHLLQP
Mvan_0803|M.vanbaalenii_PYR-1 GIKLTISDLKFMRFIGTEIFPGGQLPAVEDIEKLAADSGFTLERAHLLRP
MAB_4088c|M.abscessus_ATCC_199 GIAITISDLKFMKFIGEEIFPGGQLPAVEDIEKLAADSGFNLERVHLLQP
MUL_4538|M.ulcerans_Agy99 GVKVTMSDVRFMRFIGQEIFPGGQLPAQEDIFNFAQAAGFSVERAQLLQQ
TH_3763|M.thermoresistible__bu GIKITISDLKFMQFIGREIFPGGELPAEGDVVEFATGAGFSVERIQLLRP
Mflv_1800|M.gilvum_PYR-GCK GIPIVMSDLKFMRFIAKEIFPGGSVPGEENIVELSAEAGFTLEQTQFLNK
*: :.:.*::*::*: .*****.: . :: . : :** :*: : :.
Mb0664c|M.bovis_AF2122/97 HYARTLDAWAANLQAARERAIAVQSEEVYNNFMHYLTGCAERFRRGLINV
Rv0645c|M.tuberculosis_H37Rv HYARTLDAWAANLQAARERAIAVQSEEVYNNFMHYLTGCAERFRRGLINV
MLBr_01900|M.leprae_Br4923 HYARTLDMWATNLAAARDHALAIQPEEIYDNFMHYLTGCADRFRRGLINV
MMAR_0980|M.marinum_M HYAHTLDIWAANLAAARDRAIAVQSEQVYDNFMHYLTGCAERFRRGLVNV
MAV_4516|M.avium_104 HYARTLDTWAANLEANREQAIAIQSQEVYDRFMRYLTGCAELFRKGISNV
MSMEG_0913|M.smegmatis_MC2_155 HYARTLDMWAANLEAKKDEAIAMQGQEVYDRYMKYLTGCADFFRRGITNV
Mvan_0803|M.vanbaalenii_PYR-1 HYARTLDMWAANLEANRQEATRITSPEVYERYMHYLTGCADFFRRGITNV
MAB_4088c|M.abscessus_ATCC_199 HYAKTLDIWAQNLEQRREEAIAIQSQEVYDRFMKYLTGCADFFRRGVTNV
MUL_4538|M.ulcerans_Agy99 HYARTLNIWAANLEARKEEAIAMQSQEIYDKYMHYLTGCENFFRKGISNV
TH_3763|M.thermoresistible__bu HYARTLDIWAENLAANREPAIAMQSEEVYDRYYRYLTGCADFFRRGITNV
Mflv_1800|M.gilvum_PYR-GCK DYARTLDTWAEALEAHHDEAVAATSEEVYQRYMKYLTGCSDFFTRGISEL
.**:**: ** * :: * ::*:.: :***** : * :*: ::
Mb0664c|M.bovis_AF2122/97 AQFTMTK-
Rv0645c|M.tuberculosis_H37Rv AQFTMTK-
MLBr_01900|M.leprae_Br4923 AQFTLTK-
MMAR_0980|M.marinum_M GQFTLTK-
MAV_4516|M.avium_104 AQYTLTK-
MSMEG_0913|M.smegmatis_MC2_155 GQFTLVK-
Mvan_0803|M.vanbaalenii_PYR-1 GQFTLVKG
MAB_4088c|M.abscessus_ATCC_199 GQFTLVK-
MUL_4538|M.ulcerans_Agy99 GQFTLVK-
TH_3763|M.thermoresistible__bu GQFTLTKG
Mflv_1800|M.gilvum_PYR-GCK GQFTLVKA
.*:*:.*