For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NTAHSPDGSGRTGTARVKRGMAEMLKGGVIMDVVTPEQAKIAEGAGAVAVMALERVPADIRAQGGVSRMS DPDMIEGIISAVTIPVMAKARIGHFVEAQILQSLGVDYIDESEVLTPADYTHHIDKWKFTVPFVCGATNL GEALRRINEGAAMIRSKGEAGTGDVSNATTHMRAIGGEIRRLMSLSEDELYVAAKELQAPYELVVEVARA GKLPVTLFTAGGIATPADAAMMMQLGAEGVFVGSGIFKSGDPAQRAAAIVKATTFYDDPDVLAKVSRGLG EAMVGINVEQIAQPERLAERGW*
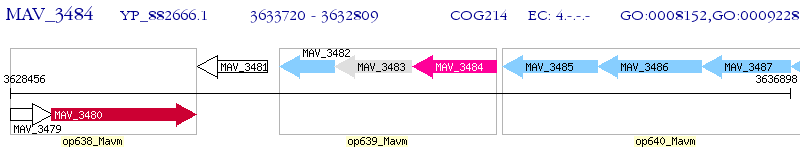
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_3484 | - | - | 100% (303) | pyridoxine biosynthesis protein |
| M. avium 104 | MAV_4744 | thiG | 1e-06 | 41.54% (65) | thiazole synthase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2638c | - | 1e-152 | 91.58% (297) | pyridoxal biosynthesis lyase PdxS |
| M. gilvum PYR-GCK | Mflv_3832 | - | 1e-155 | 94.24% (295) | pyridoxine biosynthesis protein |
| M. tuberculosis H37Rv | Rv2606c | - | 1e-152 | 91.58% (297) | pyridoxal biosynthesis lyase PdxS |
| M. leprae Br4923 | MLBr_00450 | - | 1e-154 | 93.17% (293) | pyridoxal biosynthesis lyase PdxS |
| M. abscessus ATCC 19977 | MAB_2892c | - | 1e-155 | 95.88% (291) | pyridoxal biosynthesis lyase PdxS |
| M. marinum M | MMAR_2094 | snzP | 1e-149 | 94.68% (282) | pyridoxine biosynthesis protein, SnzP |
| M. smegmatis MC2 155 | MSMEG_2937 | - | 1e-157 | 93.07% (303) | pyridoxine biosynthesis protein |
| M. thermoresistible (build 8) | TH_2055 | - | 1e-149 | 89.44% (303) | Possible pyridoxine biosynthesis protein |
| M. ulcerans Agy99 | MUL_3250 | snzP | 1e-156 | 92.38% (302) | pyridoxal biosynthesis lyase PdxS |
| M. vanbaalenii PYR-1 | Mvan_2564 | - | 1e-153 | 92.88% (295) | pyridoxine biosynthesis protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2638c|M.bovis_AF2122/97 -----------------------MDPAGNP------------------AT
Rv2606c|M.tuberculosis_H37Rv -----------------------MDPAGNP------------------AT
MMAR_2094|M.marinum_M --------------------------------------------------
MUL_3250|M.ulcerans_Agy99 -----------------------MDSATQPRGDRPDPASSKQAQSGSSQT
TH_2055|M.thermoresistible__bu --------------------VEEMTVETRDKAVGPNGSADG------AQT
Mflv_3832|M.gilvum_PYR-GCK -----MGSQARRSGSNGFAAASKLDQQRIK--EIAVESELTGAASGAPAR
Mvan_2564|M.vanbaalenii_PYR-1 -----------------------------------MESETSGAGLGAPAR
MAB_2892c|M.abscessus_ATCC_199 MRQSKSGEARHRTSRHKIAGAAALDWSQTPRFRSPQLTSTTNGTSPETGT
MSMEG_2937|M.smegmatis_MC2_155 -----------------------------------METAAQNGSSNQTGT
MAV_3484|M.avium_104 -----------------------MNTAHSP----------DGS----GRT
MLBr_00450|M.leprae_Br4923 -----------------------MDSAAQS----------NQAQSVFGQT
Mb2638c|M.bovis_AF2122/97 GTARVKRGMAEMLKGGVIMDVVTPEQARIAEGAGAVAVMALERVPADIRA
Rv2606c|M.tuberculosis_H37Rv GTARVKRGMAEMLKGGVIMDVVTPEQARIAEGAGAVAVMALERVPADIRA
MMAR_2094|M.marinum_M --------MAEMLKGGVIMDVVTPEQARIAEGAGAVAVMALERVPADIRA
MUL_3250|M.ulcerans_Agy99 GTARVKRGMAEMLKGGVIMDVVTPEQARIAEGAGAVAVMALERVPADIRA
TH_2055|M.thermoresistible__bu GTARVKRGMAEMLKGGVIMDVVTPEQARIAEGAGAVAVMALERVPADIRA
Mflv_3832|M.gilvum_PYR-GCK GTARVKRGMAEMLKGGVIMDVVTPEQARIAEGAGAVAVMALERVPADIRA
Mvan_2564|M.vanbaalenii_PYR-1 GTARVKRGMAEMLKGGVIMDVVTPEQARIAEGAGAVAVMALERVPADIRA
MAB_2892c|M.abscessus_ATCC_199 GTARVKRGMAEMLKGGVIMDVVTPEQAKIAEDAGAVAVMALERVPADIRA
MSMEG_2937|M.smegmatis_MC2_155 --ARVKRGMAEMLKGGVIMDVVTPEQARIAEAAGAVAVMALERVPADIRA
MAV_3484|M.avium_104 GTARVKRGMAEMLKGGVIMDVVTPEQAKIAEGAGAVAVMALERVPADIRA
MLBr_00450|M.leprae_Br4923 GTARVKRGMAEMLKGGVIMDVVIPEQARIAEGSGAVAVMALERVPSDIRA
************** ****:*** :************:****
Mb2638c|M.bovis_AF2122/97 QGGVSRMSDPDMIEGIIAAVTIPVMAKVRIGHFVEAQILQTLGVDYIDES
Rv2606c|M.tuberculosis_H37Rv QGGVSRMSDPDMIEGIIAAVTIPVMAKVRIGHFVEAQILQTLGVDYIDES
MMAR_2094|M.marinum_M QGGVSRMSDPDMIEGIIEAVTIPVMAKARIGHFVEAQILQSLGVDYIDES
MUL_3250|M.ulcerans_Agy99 QGGVSRMSDPDMIEGIIEAVTIPVMAKARIGHFVEAQILQSLGVDYIDES
TH_2055|M.thermoresistible__bu QGGVARMSDPDLIEGIINAVTIPVMAKVRIGHFVEAQILQSLGVDYIDES
Mflv_3832|M.gilvum_PYR-GCK QGGVSRMSDPDMIEGIIDAVTIPVMAKARIGHFVEAQILQSLGVDYVDES
Mvan_2564|M.vanbaalenii_PYR-1 QGGVSRMSDPDMIEGIIDAVTIPVMAKARIGHFVEAQILQSLGVDYIDES
MAB_2892c|M.abscessus_ATCC_199 QGGVSRMSDPDMIDGIISAVSIPVMAKARIGHFVEAQILQSLGVDYIDES
MSMEG_2937|M.smegmatis_MC2_155 QGGVSRMSDPDMIEGIIDAVTIPVMAKARIGHFVEAQILQSLGVDYVDES
MAV_3484|M.avium_104 QGGVSRMSDPDMIEGIISAVTIPVMAKARIGHFVEAQILQSLGVDYIDES
MLBr_00450|M.leprae_Br4923 QGGVSRMSDPDMIESIIAAVTIPVMAKARIGHFVEAQILQSLGVDYIDES
****:******:*:.** **:******.************:*****:***
Mb2638c|M.bovis_AF2122/97 EVLTPADYAHHIDKWNFTVPFVCGATNLGEALRRISEGAAMIRSKGEAGT
Rv2606c|M.tuberculosis_H37Rv EVLTPADYAHHIDKWNFTVPFVCGATNLGEALRRISEGAAMIRSKGEAGT
MMAR_2094|M.marinum_M EVLTPADYTHHIDKWKFTVPFVCGATNLGEALRRIGEGAAMIRSKGEAGT
MUL_3250|M.ulcerans_Agy99 EVLTPADYTHHIDKWKFTVPFVCGATNLGEALRRIGEGAAMIRSKGEAGT
TH_2055|M.thermoresistible__bu EVLTPADYTHHIDKWKFTVPFVCGATNLGEALRRIAEGAAMIRSKGEAGT
Mflv_3832|M.gilvum_PYR-GCK EVLTPADYTNHIDKWKFTVPFVCGATNLGEALRRITEGAAMIRSKGEAGT
Mvan_2564|M.vanbaalenii_PYR-1 EVLTPADYANHIDKWRFTVPFVCGATNLGEALRRLTEGAAMIRSKGEAGT
MAB_2892c|M.abscessus_ATCC_199 EVLTPADYTNHIDKWKFTVPFVCGATNLGEALRRITEGAAMIRSKGEAGT
MSMEG_2937|M.smegmatis_MC2_155 EVLTPADYTNHIDKWKFTVPFVCGATNLGEALRRITEGAAMIRSKGEAGT
MAV_3484|M.avium_104 EVLTPADYTHHIDKWKFTVPFVCGATNLGEALRRINEGAAMIRSKGEAGT
MLBr_00450|M.leprae_Br4923 EVLTPADYTHHIDKWKFTVPFVCGATNLGEALRRINEGAAMIRSKGEAGT
********::*****.******************: **************
Mb2638c|M.bovis_AF2122/97 GDVSNATTHMRAIGGEIRRLTSMSEDELFVAAKELQAPYELVAEVARAGK
Rv2606c|M.tuberculosis_H37Rv GDVSNATTHMRAIGGEIRRLTSMSEDELFVAAKELQAPYELVAEVARAGK
MMAR_2094|M.marinum_M GDVSNATTHMRSIAGEIRRLTSMSEDELFVAAKELQAPYELVAEVARAGK
MUL_3250|M.ulcerans_Agy99 GDVSNATTHMRSIAGEIRRLTSMSEDELFVAAKELQAPYELVVEVARAGK
TH_2055|M.thermoresistible__bu GDVSNATTHMRQIRAEIRRLSALPEDELYVAAKELQAPYELVAEVARTGK
Mflv_3832|M.gilvum_PYR-GCK GDVSNATTHMRKIGGEIRRLTSLSEDELYVAAKELQAPYDLVVEVAQAGK
Mvan_2564|M.vanbaalenii_PYR-1 GDVSNATTHMRKIGGEIRRLTSLSEDELYVAAKELQAPYDLVVEVARAGK
MAB_2892c|M.abscessus_ATCC_199 GDVSNATTHMRKIGGEIRRLTSLAEDELYVAAKELQAPYELVVEVARAGK
MSMEG_2937|M.smegmatis_MC2_155 GDVSNATTHMRKIGGEIRRLTSMSEDELYVAAKELQAPYELVVEVARAGK
MAV_3484|M.avium_104 GDVSNATTHMRAIGGEIRRLMSLSEDELYVAAKELQAPYELVVEVARAGK
MLBr_00450|M.leprae_Br4923 GDVSNATTHMRAIAGDIRRLTSLSEDELYVAAKELHAPYELVIEVARTNK
*********** * .:**** ::.****:******:***:** ***::.*
Mb2638c|M.bovis_AF2122/97 LPVTLFTAGGIATPADAAMMMQLGAEGVFVGSGIFKSGAPEHRAAAIVKA
Rv2606c|M.tuberculosis_H37Rv LPVTLFTAGGIATPADAAMMMQLGAEGVFVGSGIFKSGAPEHRAAAIVKA
MMAR_2094|M.marinum_M LPVTLFTAGGIATPADAAMMMQLGAEGVFVGSGIFKSGAPEHRAAAIVKA
MUL_3250|M.ulcerans_Agy99 LPVTLFTAGGIATPADAAMMMQLGAEGVFVGSGIFKSGAPEHRAAAIVKA
TH_2055|M.thermoresistible__bu LPVTLFTAGGIATPADAAMMMQLGAEGVFVGSGIFKSGNPEQRAAAIVKA
Mflv_3832|M.gilvum_PYR-GCK LPVTLFTAGGIATPADAAMMMQLGAEGVFVGSGIFKSGNPAERAAAIVKA
Mvan_2564|M.vanbaalenii_PYR-1 LPVTLFTAGGIATPADAAMMMQLGAEGVFVGSGIFKSGNPAERAAAIVKA
MAB_2892c|M.abscessus_ATCC_199 LPVTLFTAGGIATPADAAMMMQLGAEGVFVGSGIFKSGNPEQRAAAIVKA
MSMEG_2937|M.smegmatis_MC2_155 LPVTLFTAGGIATPADAAMMMQLGAEGVFVGSGIFKSGNPEQRAAAIVKA
MAV_3484|M.avium_104 LPVTLFTAGGIATPADAAMMMQLGAEGVFVGSGIFKSGDPAQRAAAIVKA
MLBr_00450|M.leprae_Br4923 LPVTLFTAGGIATPADAAMMMQLGAEGIFVGSGIFKSSDPAQRAAAIVKA
***************************:*********. * .********
Mb2638c|M.bovis_AF2122/97 TTFFDDPDVLAKVSRGLGEAMVGINVDEIAVGHRLAQRGW
Rv2606c|M.tuberculosis_H37Rv TTFFDDPDVLAKVSRGLGEAMVGINVDEIAVGHRLAQRGW
MMAR_2094|M.marinum_M TTFYDDPDVLAKVSRGLGEAMVGINVEEIAQPHRLAQRGW
MUL_3250|M.ulcerans_Agy99 TTFYDDPDVLAKVSRGLGEAMVGINVEEIAQPHRLAQRGW
TH_2055|M.thermoresistible__bu TTFYDDPDVLAKVSRNLGEPMVGINVADVPVPHRLAERGW
Mflv_3832|M.gilvum_PYR-GCK TTFYDDPDVLAKVSRGLGEAMVGINVEDIAQPHRLAERGW
Mvan_2564|M.vanbaalenii_PYR-1 TTFYDDPDVLAKVSRGLGEAMVGINVDDIPVPHRLAERGW
MAB_2892c|M.abscessus_ATCC_199 TTFYDDPDVLAKVSRGLGEAMVGINVEDIAQPHRLAERGW
MSMEG_2937|M.smegmatis_MC2_155 TTFYDDPDVLAKVSRGLGEAMVGINVEEIAQPHRLAERGW
MAV_3484|M.avium_104 TTFYDDPDVLAKVSRGLGEAMVGINVEQIAQPERLAERGW
MLBr_00450|M.leprae_Br4923 TTFYDDPDVLAKVSRGLGEAMAGIDVEQIAQPDRLAQRGW
***:***********.***.*.**:* ::. .***:***