For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VESETSGAGLGAPARGTARVKRGMAEMLKGGVIMDVVTPEQARIAEGAGAVAVMALERVPADIRAQGGVS RMSDPDMIEGIIDAVTIPVMAKARIGHFVEAQILQSLGVDYIDESEVLTPADYANHIDKWRFTVPFVCGA TNLGEALRRLTEGAAMIRSKGEAGTGDVSNATTHMRKIGGEIRRLTSLSEDELYVAAKELQAPYDLVVEV ARAGKLPVTLFTAGGIATPADAAMMMQLGAEGVFVGSGIFKSGNPAERAAAIVKATTFYDDPDVLAKVSR GLGEAMVGINVDDIPVPHRLAERGW
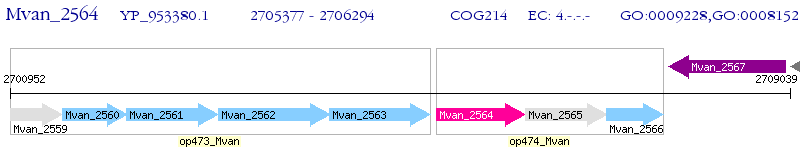
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_2564 | - | - | 100% (305) | pyridoxine biosynthesis protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2638c | - | 1e-153 | 92.54% (295) | pyridoxal biosynthesis lyase PdxS |
| M. gilvum PYR-GCK | Mflv_3832 | - | 1e-165 | 96.07% (305) | pyridoxine biosynthesis protein |
| M. tuberculosis H37Rv | Rv2606c | - | 1e-153 | 92.54% (295) | pyridoxal biosynthesis lyase PdxS |
| M. leprae Br4923 | MLBr_00450 | - | 1e-146 | 89.66% (290) | pyridoxal biosynthesis lyase PdxS |
| M. abscessus ATCC 19977 | MAB_2892c | - | 1e-155 | 94.83% (290) | pyridoxal biosynthesis lyase PdxS |
| M. marinum M | MMAR_2094 | snzP | 1e-147 | 92.91% (282) | pyridoxine biosynthesis protein, SnzP |
| M. avium 104 | MAV_3484 | - | 1e-153 | 92.88% (295) | pyridoxine biosynthesis protein |
| M. smegmatis MC2 155 | MSMEG_2937 | - | 1e-156 | 92.38% (302) | pyridoxine biosynthesis protein |
| M. thermoresistible (build 8) | TH_2055 | - | 1e-150 | 89.67% (300) | Possible pyridoxine biosynthesis protein |
| M. ulcerans Agy99 | MUL_3250 | snzP | 1e-152 | 92.20% (295) | pyridoxal biosynthesis lyase PdxS |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2638c|M.bovis_AF2122/97 -----------------------------------MDPAGNP--------
Rv2606c|M.tuberculosis_H37Rv -----------------------------------MDPAGNP--------
MMAR_2094|M.marinum_M --------------------------------------------------
MUL_3250|M.ulcerans_Agy99 -----------------------------------MDSATQPRGDRPDPA
TH_2055|M.thermoresistible__bu --------------------------------VEEMTVETRDKAVGPNGS
MLBr_00450|M.leprae_Br4923 -----------------------------------MDSAAQSNQAQSVF-
MAV_3484|M.avium_104 -----------------------------------MNTAHSPDGS-----
Mvan_2564|M.vanbaalenii_PYR-1 -----------------------------------MESETSGAGLGA---
Mflv_3832|M.gilvum_PYR-GCK -----MGSQARRSGSNGFAAASKLDQQRIK--EIAVESELTGAASGA---
MAB_2892c|M.abscessus_ATCC_199 MRQSKSGEARHRTSRHKIAGAAALDWSQTPRFRSPQLTSTTNGTSPE---
MSMEG_2937|M.smegmatis_MC2_155 -----------------------------------METAAQNGSSNQ---
Mb2638c|M.bovis_AF2122/97 ----------ATGTARVKRGMAEMLKGGVIMDVVTPEQARIAEGAGAVAV
Rv2606c|M.tuberculosis_H37Rv ----------ATGTARVKRGMAEMLKGGVIMDVVTPEQARIAEGAGAVAV
MMAR_2094|M.marinum_M --------------------MAEMLKGGVIMDVVTPEQARIAEGAGAVAV
MUL_3250|M.ulcerans_Agy99 SSKQAQSGSSQTGTARVKRGMAEMLKGGVIMDVVTPEQARIAEGAGAVAV
TH_2055|M.thermoresistible__bu ADG------AQTGTARVKRGMAEMLKGGVIMDVVTPEQARIAEGAGAVAV
MLBr_00450|M.leprae_Br4923 ---------GQTGTARVKRGMAEMLKGGVIMDVVIPEQARIAEGSGAVAV
MAV_3484|M.avium_104 ---------GRTGTARVKRGMAEMLKGGVIMDVVTPEQAKIAEGAGAVAV
Mvan_2564|M.vanbaalenii_PYR-1 ---------PARGTARVKRGMAEMLKGGVIMDVVTPEQARIAEGAGAVAV
Mflv_3832|M.gilvum_PYR-GCK ---------PARGTARVKRGMAEMLKGGVIMDVVTPEQARIAEGAGAVAV
MAB_2892c|M.abscessus_ATCC_199 ---------TGTGTARVKRGMAEMLKGGVIMDVVTPEQAKIAEDAGAVAV
MSMEG_2937|M.smegmatis_MC2_155 ---------TGT--ARVKRGMAEMLKGGVIMDVVTPEQARIAEAAGAVAV
************** ****:*** :*****
Mb2638c|M.bovis_AF2122/97 MALERVPADIRAQGGVSRMSDPDMIEGIIAAVTIPVMAKVRIGHFVEAQI
Rv2606c|M.tuberculosis_H37Rv MALERVPADIRAQGGVSRMSDPDMIEGIIAAVTIPVMAKVRIGHFVEAQI
MMAR_2094|M.marinum_M MALERVPADIRAQGGVSRMSDPDMIEGIIEAVTIPVMAKARIGHFVEAQI
MUL_3250|M.ulcerans_Agy99 MALERVPADIRAQGGVSRMSDPDMIEGIIEAVTIPVMAKARIGHFVEAQI
TH_2055|M.thermoresistible__bu MALERVPADIRAQGGVARMSDPDLIEGIINAVTIPVMAKVRIGHFVEAQI
MLBr_00450|M.leprae_Br4923 MALERVPSDIRAQGGVSRMSDPDMIESIIAAVTIPVMAKARIGHFVEAQI
MAV_3484|M.avium_104 MALERVPADIRAQGGVSRMSDPDMIEGIISAVTIPVMAKARIGHFVEAQI
Mvan_2564|M.vanbaalenii_PYR-1 MALERVPADIRAQGGVSRMSDPDMIEGIIDAVTIPVMAKARIGHFVEAQI
Mflv_3832|M.gilvum_PYR-GCK MALERVPADIRAQGGVSRMSDPDMIEGIIDAVTIPVMAKARIGHFVEAQI
MAB_2892c|M.abscessus_ATCC_199 MALERVPADIRAQGGVSRMSDPDMIDGIISAVSIPVMAKARIGHFVEAQI
MSMEG_2937|M.smegmatis_MC2_155 MALERVPADIRAQGGVSRMSDPDMIEGIIDAVTIPVMAKARIGHFVEAQI
*******:********:******:*:.** **:******.**********
Mb2638c|M.bovis_AF2122/97 LQTLGVDYIDESEVLTPADYAHHIDKWNFTVPFVCGATNLGEALRRISEG
Rv2606c|M.tuberculosis_H37Rv LQTLGVDYIDESEVLTPADYAHHIDKWNFTVPFVCGATNLGEALRRISEG
MMAR_2094|M.marinum_M LQSLGVDYIDESEVLTPADYTHHIDKWKFTVPFVCGATNLGEALRRIGEG
MUL_3250|M.ulcerans_Agy99 LQSLGVDYIDESEVLTPADYTHHIDKWKFTVPFVCGATNLGEALRRIGEG
TH_2055|M.thermoresistible__bu LQSLGVDYIDESEVLTPADYTHHIDKWKFTVPFVCGATNLGEALRRIAEG
MLBr_00450|M.leprae_Br4923 LQSLGVDYIDESEVLTPADYTHHIDKWKFTVPFVCGATNLGEALRRINEG
MAV_3484|M.avium_104 LQSLGVDYIDESEVLTPADYTHHIDKWKFTVPFVCGATNLGEALRRINEG
Mvan_2564|M.vanbaalenii_PYR-1 LQSLGVDYIDESEVLTPADYANHIDKWRFTVPFVCGATNLGEALRRLTEG
Mflv_3832|M.gilvum_PYR-GCK LQSLGVDYVDESEVLTPADYTNHIDKWKFTVPFVCGATNLGEALRRITEG
MAB_2892c|M.abscessus_ATCC_199 LQSLGVDYIDESEVLTPADYTNHIDKWKFTVPFVCGATNLGEALRRITEG
MSMEG_2937|M.smegmatis_MC2_155 LQSLGVDYVDESEVLTPADYTNHIDKWKFTVPFVCGATNLGEALRRITEG
**:*****:***********::*****.******************: **
Mb2638c|M.bovis_AF2122/97 AAMIRSKGEAGTGDVSNATTHMRAIGGEIRRLTSMSEDELFVAAKELQAP
Rv2606c|M.tuberculosis_H37Rv AAMIRSKGEAGTGDVSNATTHMRAIGGEIRRLTSMSEDELFVAAKELQAP
MMAR_2094|M.marinum_M AAMIRSKGEAGTGDVSNATTHMRSIAGEIRRLTSMSEDELFVAAKELQAP
MUL_3250|M.ulcerans_Agy99 AAMIRSKGEAGTGDVSNATTHMRSIAGEIRRLTSMSEDELFVAAKELQAP
TH_2055|M.thermoresistible__bu AAMIRSKGEAGTGDVSNATTHMRQIRAEIRRLSALPEDELYVAAKELQAP
MLBr_00450|M.leprae_Br4923 AAMIRSKGEAGTGDVSNATTHMRAIAGDIRRLTSLSEDELYVAAKELHAP
MAV_3484|M.avium_104 AAMIRSKGEAGTGDVSNATTHMRAIGGEIRRLMSLSEDELYVAAKELQAP
Mvan_2564|M.vanbaalenii_PYR-1 AAMIRSKGEAGTGDVSNATTHMRKIGGEIRRLTSLSEDELYVAAKELQAP
Mflv_3832|M.gilvum_PYR-GCK AAMIRSKGEAGTGDVSNATTHMRKIGGEIRRLTSLSEDELYVAAKELQAP
MAB_2892c|M.abscessus_ATCC_199 AAMIRSKGEAGTGDVSNATTHMRKIGGEIRRLTSLAEDELYVAAKELQAP
MSMEG_2937|M.smegmatis_MC2_155 AAMIRSKGEAGTGDVSNATTHMRKIGGEIRRLTSMSEDELYVAAKELQAP
*********************** * .:**** ::.****:******:**
Mb2638c|M.bovis_AF2122/97 YELVAEVARAGKLPVTLFTAGGIATPADAAMMMQLGAEGVFVGSGIFKSG
Rv2606c|M.tuberculosis_H37Rv YELVAEVARAGKLPVTLFTAGGIATPADAAMMMQLGAEGVFVGSGIFKSG
MMAR_2094|M.marinum_M YELVAEVARAGKLPVTLFTAGGIATPADAAMMMQLGAEGVFVGSGIFKSG
MUL_3250|M.ulcerans_Agy99 YELVVEVARAGKLPVTLFTAGGIATPADAAMMMQLGAEGVFVGSGIFKSG
TH_2055|M.thermoresistible__bu YELVAEVARTGKLPVTLFTAGGIATPADAAMMMQLGAEGVFVGSGIFKSG
MLBr_00450|M.leprae_Br4923 YELVIEVARTNKLPVTLFTAGGIATPADAAMMMQLGAEGIFVGSGIFKSS
MAV_3484|M.avium_104 YELVVEVARAGKLPVTLFTAGGIATPADAAMMMQLGAEGVFVGSGIFKSG
Mvan_2564|M.vanbaalenii_PYR-1 YDLVVEVARAGKLPVTLFTAGGIATPADAAMMMQLGAEGVFVGSGIFKSG
Mflv_3832|M.gilvum_PYR-GCK YDLVVEVAQAGKLPVTLFTAGGIATPADAAMMMQLGAEGVFVGSGIFKSG
MAB_2892c|M.abscessus_ATCC_199 YELVVEVARAGKLPVTLFTAGGIATPADAAMMMQLGAEGVFVGSGIFKSG
MSMEG_2937|M.smegmatis_MC2_155 YELVVEVARAGKLPVTLFTAGGIATPADAAMMMQLGAEGVFVGSGIFKSG
*:** ***::.****************************:*********.
Mb2638c|M.bovis_AF2122/97 APEHRAAAIVKATTFFDDPDVLAKVSRGLGEAMVGINVDEIAVGHRLAQR
Rv2606c|M.tuberculosis_H37Rv APEHRAAAIVKATTFFDDPDVLAKVSRGLGEAMVGINVDEIAVGHRLAQR
MMAR_2094|M.marinum_M APEHRAAAIVKATTFYDDPDVLAKVSRGLGEAMVGINVEEIAQPHRLAQR
MUL_3250|M.ulcerans_Agy99 APEHRAAAIVKATTFYDDPDVLAKVSRGLGEAMVGINVEEIAQPHRLAQR
TH_2055|M.thermoresistible__bu NPEQRAAAIVKATTFYDDPDVLAKVSRNLGEPMVGINVADVPVPHRLAER
MLBr_00450|M.leprae_Br4923 DPAQRAAAIVKATTFYDDPDVLAKVSRGLGEAMAGIDVEQIAQPDRLAQR
MAV_3484|M.avium_104 DPAQRAAAIVKATTFYDDPDVLAKVSRGLGEAMVGINVEQIAQPERLAER
Mvan_2564|M.vanbaalenii_PYR-1 NPAERAAAIVKATTFYDDPDVLAKVSRGLGEAMVGINVDDIPVPHRLAER
Mflv_3832|M.gilvum_PYR-GCK NPAERAAAIVKATTFYDDPDVLAKVSRGLGEAMVGINVEDIAQPHRLAER
MAB_2892c|M.abscessus_ATCC_199 NPEQRAAAIVKATTFYDDPDVLAKVSRGLGEAMVGINVEDIAQPHRLAER
MSMEG_2937|M.smegmatis_MC2_155 NPEQRAAAIVKATTFYDDPDVLAKVSRGLGEAMVGINVEEIAQPHRLAER
* .***********:***********.***.*.**:* ::. .***:*
Mb2638c|M.bovis_AF2122/97 GW
Rv2606c|M.tuberculosis_H37Rv GW
MMAR_2094|M.marinum_M GW
MUL_3250|M.ulcerans_Agy99 GW
TH_2055|M.thermoresistible__bu GW
MLBr_00450|M.leprae_Br4923 GW
MAV_3484|M.avium_104 GW
Mvan_2564|M.vanbaalenii_PYR-1 GW
Mflv_3832|M.gilvum_PYR-GCK GW
MAB_2892c|M.abscessus_ATCC_199 GW
MSMEG_2937|M.smegmatis_MC2_155 GW
**