For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RQSKSGEARHRTSRHKIAGAAALDWSQTPRFRSPQLTSTTNGTSPETGTGTARVKRGMAEMLKGGVIMDV VTPEQAKIAEDAGAVAVMALERVPADIRAQGGVSRMSDPDMIDGIISAVSIPVMAKARIGHFVEAQILQS LGVDYIDESEVLTPADYTNHIDKWKFTVPFVCGATNLGEALRRITEGAAMIRSKGEAGTGDVSNATTHMR KIGGEIRRLTSLAEDELYVAAKELQAPYELVVEVARAGKLPVTLFTAGGIATPADAAMMMQLGAEGVFVG SGIFKSGNPEQRAAAIVKATTFYDDPDVLAKVSRGLGEAMVGINVEDIAQPHRLAERGW*
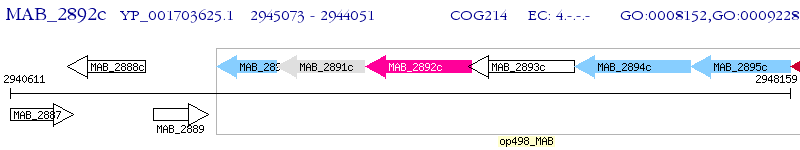
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2892c | - | - | 100% (340) | pyridoxal biosynthesis lyase PdxS |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2638c | - | 1e-150 | 91.75% (291) | pyridoxal biosynthesis lyase PdxS |
| M. gilvum PYR-GCK | Mflv_3832 | - | 1e-158 | 85.97% (335) | pyridoxine biosynthesis protein |
| M. tuberculosis H37Rv | Rv2606c | - | 1e-150 | 91.75% (291) | pyridoxal biosynthesis lyase PdxS |
| M. leprae Br4923 | MLBr_00450 | - | 1e-148 | 90.38% (291) | pyridoxal biosynthesis lyase PdxS |
| M. marinum M | MMAR_2094 | snzP | 1e-148 | 93.97% (282) | pyridoxine biosynthesis protein, SnzP |
| M. avium 104 | MAV_3484 | - | 1e-155 | 95.88% (291) | pyridoxine biosynthesis protein |
| M. smegmatis MC2 155 | MSMEG_2937 | - | 1e-160 | 94.39% (303) | pyridoxine biosynthesis protein |
| M. thermoresistible (build 8) | TH_2055 | - | 1e-151 | 89.97% (299) | Possible pyridoxine biosynthesis protein |
| M. ulcerans Agy99 | MUL_3250 | snzP | 1e-156 | 88.64% (317) | pyridoxal biosynthesis lyase PdxS |
| M. vanbaalenii PYR-1 | Mvan_2564 | - | 1e-155 | 94.83% (290) | pyridoxine biosynthesis protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2638c|M.bovis_AF2122/97 ---------------------------------MDPAGNP----------
Rv2606c|M.tuberculosis_H37Rv ---------------------------------MDPAGNP----------
MMAR_2094|M.marinum_M --------------------------------------------------
MUL_3250|M.ulcerans_Agy99 ---------------------------------MDSATQPRGDRPDPASS
TH_2055|M.thermoresistible__bu ------------------------------VEEMTVETRDKAVGPNGSAD
MLBr_00450|M.leprae_Br4923 ---------------------------------MDSAAQSNQAQSVF---
MAV_3484|M.avium_104 ---------------------------------MNTAHSPDGS-------
MAB_2892c|M.abscessus_ATCC_199 MRQSKSGEARHRTSRHKIAGAAALDWSQTPRFRSPQLTSTTNGTSPE---
MSMEG_2937|M.smegmatis_MC2_155 -----------------------------------METAAQNGSSNQ---
Mflv_3832|M.gilvum_PYR-GCK -----MGSQARRSGSNGFAAASKLDQQRIKEIAVESELTGAASGAPA---
Mvan_2564|M.vanbaalenii_PYR-1 ---------------------------------MESETSGAGLGAPA---
Mb2638c|M.bovis_AF2122/97 --------ATGTARVKRGMAEMLKGGVIMDVVTPEQARIAEGAGAVAVMA
Rv2606c|M.tuberculosis_H37Rv --------ATGTARVKRGMAEMLKGGVIMDVVTPEQARIAEGAGAVAVMA
MMAR_2094|M.marinum_M ------------------MAEMLKGGVIMDVVTPEQARIAEGAGAVAVMA
MUL_3250|M.ulcerans_Agy99 KQAQSGSSQTGTARVKRGMAEMLKGGVIMDVVTPEQARIAEGAGAVAVMA
TH_2055|M.thermoresistible__bu G------AQTGTARVKRGMAEMLKGGVIMDVVTPEQARIAEGAGAVAVMA
MLBr_00450|M.leprae_Br4923 -------GQTGTARVKRGMAEMLKGGVIMDVVIPEQARIAEGSGAVAVMA
MAV_3484|M.avium_104 -------GRTGTARVKRGMAEMLKGGVIMDVVTPEQAKIAEGAGAVAVMA
MAB_2892c|M.abscessus_ATCC_199 -------TGTGTARVKRGMAEMLKGGVIMDVVTPEQAKIAEDAGAVAVMA
MSMEG_2937|M.smegmatis_MC2_155 -------TGT--ARVKRGMAEMLKGGVIMDVVTPEQARIAEAAGAVAVMA
Mflv_3832|M.gilvum_PYR-GCK -------RGT--ARVKRGMAEMLKGGVIMDVVTPEQARIAEGAGAVAVMA
Mvan_2564|M.vanbaalenii_PYR-1 -------RGT--ARVKRGMAEMLKGGVIMDVVTPEQARIAEGAGAVAVMA
************** ****:*** :*******
Mb2638c|M.bovis_AF2122/97 LERVPADIRAQGGVSRMSDPDMIEGIIAAVTIPVMAKVRIGHFVEAQILQ
Rv2606c|M.tuberculosis_H37Rv LERVPADIRAQGGVSRMSDPDMIEGIIAAVTIPVMAKVRIGHFVEAQILQ
MMAR_2094|M.marinum_M LERVPADIRAQGGVSRMSDPDMIEGIIEAVTIPVMAKARIGHFVEAQILQ
MUL_3250|M.ulcerans_Agy99 LERVPADIRAQGGVSRMSDPDMIEGIIEAVTIPVMAKARIGHFVEAQILQ
TH_2055|M.thermoresistible__bu LERVPADIRAQGGVARMSDPDLIEGIINAVTIPVMAKVRIGHFVEAQILQ
MLBr_00450|M.leprae_Br4923 LERVPSDIRAQGGVSRMSDPDMIESIIAAVTIPVMAKARIGHFVEAQILQ
MAV_3484|M.avium_104 LERVPADIRAQGGVSRMSDPDMIEGIISAVTIPVMAKARIGHFVEAQILQ
MAB_2892c|M.abscessus_ATCC_199 LERVPADIRAQGGVSRMSDPDMIDGIISAVSIPVMAKARIGHFVEAQILQ
MSMEG_2937|M.smegmatis_MC2_155 LERVPADIRAQGGVSRMSDPDMIEGIIDAVTIPVMAKARIGHFVEAQILQ
Mflv_3832|M.gilvum_PYR-GCK LERVPADIRAQGGVSRMSDPDMIEGIIDAVTIPVMAKARIGHFVEAQILQ
Mvan_2564|M.vanbaalenii_PYR-1 LERVPADIRAQGGVSRMSDPDMIEGIIDAVTIPVMAKARIGHFVEAQILQ
*****:********:******:*:.** **:******.************
Mb2638c|M.bovis_AF2122/97 TLGVDYIDESEVLTPADYAHHIDKWNFTVPFVCGATNLGEALRRISEGAA
Rv2606c|M.tuberculosis_H37Rv TLGVDYIDESEVLTPADYAHHIDKWNFTVPFVCGATNLGEALRRISEGAA
MMAR_2094|M.marinum_M SLGVDYIDESEVLTPADYTHHIDKWKFTVPFVCGATNLGEALRRIGEGAA
MUL_3250|M.ulcerans_Agy99 SLGVDYIDESEVLTPADYTHHIDKWKFTVPFVCGATNLGEALRRIGEGAA
TH_2055|M.thermoresistible__bu SLGVDYIDESEVLTPADYTHHIDKWKFTVPFVCGATNLGEALRRIAEGAA
MLBr_00450|M.leprae_Br4923 SLGVDYIDESEVLTPADYTHHIDKWKFTVPFVCGATNLGEALRRINEGAA
MAV_3484|M.avium_104 SLGVDYIDESEVLTPADYTHHIDKWKFTVPFVCGATNLGEALRRINEGAA
MAB_2892c|M.abscessus_ATCC_199 SLGVDYIDESEVLTPADYTNHIDKWKFTVPFVCGATNLGEALRRITEGAA
MSMEG_2937|M.smegmatis_MC2_155 SLGVDYVDESEVLTPADYTNHIDKWKFTVPFVCGATNLGEALRRITEGAA
Mflv_3832|M.gilvum_PYR-GCK SLGVDYVDESEVLTPADYTNHIDKWKFTVPFVCGATNLGEALRRITEGAA
Mvan_2564|M.vanbaalenii_PYR-1 SLGVDYIDESEVLTPADYANHIDKWRFTVPFVCGATNLGEALRRLTEGAA
:*****:***********::*****.******************: ****
Mb2638c|M.bovis_AF2122/97 MIRSKGEAGTGDVSNATTHMRAIGGEIRRLTSMSEDELFVAAKELQAPYE
Rv2606c|M.tuberculosis_H37Rv MIRSKGEAGTGDVSNATTHMRAIGGEIRRLTSMSEDELFVAAKELQAPYE
MMAR_2094|M.marinum_M MIRSKGEAGTGDVSNATTHMRSIAGEIRRLTSMSEDELFVAAKELQAPYE
MUL_3250|M.ulcerans_Agy99 MIRSKGEAGTGDVSNATTHMRSIAGEIRRLTSMSEDELFVAAKELQAPYE
TH_2055|M.thermoresistible__bu MIRSKGEAGTGDVSNATTHMRQIRAEIRRLSALPEDELYVAAKELQAPYE
MLBr_00450|M.leprae_Br4923 MIRSKGEAGTGDVSNATTHMRAIAGDIRRLTSLSEDELYVAAKELHAPYE
MAV_3484|M.avium_104 MIRSKGEAGTGDVSNATTHMRAIGGEIRRLMSLSEDELYVAAKELQAPYE
MAB_2892c|M.abscessus_ATCC_199 MIRSKGEAGTGDVSNATTHMRKIGGEIRRLTSLAEDELYVAAKELQAPYE
MSMEG_2937|M.smegmatis_MC2_155 MIRSKGEAGTGDVSNATTHMRKIGGEIRRLTSMSEDELYVAAKELQAPYE
Mflv_3832|M.gilvum_PYR-GCK MIRSKGEAGTGDVSNATTHMRKIGGEIRRLTSLSEDELYVAAKELQAPYD
Mvan_2564|M.vanbaalenii_PYR-1 MIRSKGEAGTGDVSNATTHMRKIGGEIRRLTSLSEDELYVAAKELQAPYD
********************* * .:**** ::.****:******:***:
Mb2638c|M.bovis_AF2122/97 LVAEVARAGKLPVTLFTAGGIATPADAAMMMQLGAEGVFVGSGIFKSGAP
Rv2606c|M.tuberculosis_H37Rv LVAEVARAGKLPVTLFTAGGIATPADAAMMMQLGAEGVFVGSGIFKSGAP
MMAR_2094|M.marinum_M LVAEVARAGKLPVTLFTAGGIATPADAAMMMQLGAEGVFVGSGIFKSGAP
MUL_3250|M.ulcerans_Agy99 LVVEVARAGKLPVTLFTAGGIATPADAAMMMQLGAEGVFVGSGIFKSGAP
TH_2055|M.thermoresistible__bu LVAEVARTGKLPVTLFTAGGIATPADAAMMMQLGAEGVFVGSGIFKSGNP
MLBr_00450|M.leprae_Br4923 LVIEVARTNKLPVTLFTAGGIATPADAAMMMQLGAEGIFVGSGIFKSSDP
MAV_3484|M.avium_104 LVVEVARAGKLPVTLFTAGGIATPADAAMMMQLGAEGVFVGSGIFKSGDP
MAB_2892c|M.abscessus_ATCC_199 LVVEVARAGKLPVTLFTAGGIATPADAAMMMQLGAEGVFVGSGIFKSGNP
MSMEG_2937|M.smegmatis_MC2_155 LVVEVARAGKLPVTLFTAGGIATPADAAMMMQLGAEGVFVGSGIFKSGNP
Mflv_3832|M.gilvum_PYR-GCK LVVEVAQAGKLPVTLFTAGGIATPADAAMMMQLGAEGVFVGSGIFKSGNP
Mvan_2564|M.vanbaalenii_PYR-1 LVVEVARAGKLPVTLFTAGGIATPADAAMMMQLGAEGVFVGSGIFKSGNP
** ***::.****************************:*********. *
Mb2638c|M.bovis_AF2122/97 EHRAAAIVKATTFFDDPDVLAKVSRGLGEAMVGINVDEIAVGHRLAQRGW
Rv2606c|M.tuberculosis_H37Rv EHRAAAIVKATTFFDDPDVLAKVSRGLGEAMVGINVDEIAVGHRLAQRGW
MMAR_2094|M.marinum_M EHRAAAIVKATTFYDDPDVLAKVSRGLGEAMVGINVEEIAQPHRLAQRGW
MUL_3250|M.ulcerans_Agy99 EHRAAAIVKATTFYDDPDVLAKVSRGLGEAMVGINVEEIAQPHRLAQRGW
TH_2055|M.thermoresistible__bu EQRAAAIVKATTFYDDPDVLAKVSRNLGEPMVGINVADVPVPHRLAERGW
MLBr_00450|M.leprae_Br4923 AQRAAAIVKATTFYDDPDVLAKVSRGLGEAMAGIDVEQIAQPDRLAQRGW
MAV_3484|M.avium_104 AQRAAAIVKATTFYDDPDVLAKVSRGLGEAMVGINVEQIAQPERLAERGW
MAB_2892c|M.abscessus_ATCC_199 EQRAAAIVKATTFYDDPDVLAKVSRGLGEAMVGINVEDIAQPHRLAERGW
MSMEG_2937|M.smegmatis_MC2_155 EQRAAAIVKATTFYDDPDVLAKVSRGLGEAMVGINVEEIAQPHRLAERGW
Mflv_3832|M.gilvum_PYR-GCK AERAAAIVKATTFYDDPDVLAKVSRGLGEAMVGINVEDIAQPHRLAERGW
Mvan_2564|M.vanbaalenii_PYR-1 AERAAAIVKATTFYDDPDVLAKVSRGLGEAMVGINVDDIPVPHRLAERGW
.***********:***********.***.*.**:* ::. .***:***