For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VDSKLTIADRSFSSRLIMGTGGAGNLAVLEEALIASGTELTTVAIRRVDAEGGTGLLDLLNRLGITPLPN TAGCRGAAEAVLTAQLAREALNTNWIKLEVIADERTLLPDAVELVRAAEQLVDDGFVVLPYTNDDPVLAR RLEDAGCAAVMPLGSPIGTGLGITNPHNIEMIVAGAGVPVVLDAGIGTASDAALAMELGCDAVLLATAVT RAGDPAAMAAAMSAAVTAGYLARRAGRIPKRFWAQASSPPR*
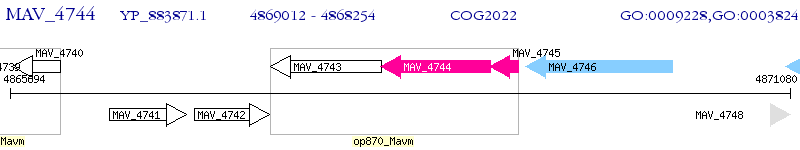
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_4744 | thiG | - | 100% (252) | thiazole synthase |
| M. avium 104 | MAV_3484 | - | 1e-06 | 41.54% (65) | pyridoxine biosynthesis protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0425 | thiG | 1e-126 | 91.67% (252) | thiazole synthase |
| M. gilvum PYR-GCK | Mflv_0195 | thiG | 1e-119 | 87.40% (246) | thiazole synthase |
| M. tuberculosis H37Rv | Rv0417 | thiG | 1e-125 | 91.27% (252) | thiazole synthase |
| M. leprae Br4923 | MLBr_00297 | thiG | 1e-119 | 86.80% (250) | thiazole synthase |
| M. abscessus ATCC 19977 | MAB_4216c | thiG | 1e-118 | 87.35% (245) | thiazole synthase |
| M. marinum M | MMAR_0720 | thiG | 1e-131 | 95.20% (250) | thiamin biosynthesis protein ThiG |
| M. smegmatis MC2 155 | MSMEG_0793 | thiG | 1e-119 | 88.00% (250) | thiazole synthase |
| M. thermoresistible (build 8) | TH_2314 | thiG | 1e-122 | 88.66% (247) | PROBABLE THIAMIN BIOSYNTHESIS PROTEIN THIG (THIAZOLE |
| M. ulcerans Agy99 | MUL_2803 | thiG | 1e-129 | 94.00% (250) | thiazole synthase |
| M. vanbaalenii PYR-1 | Mvan_0710 | thiG | 1e-120 | 88.21% (246) | thiazole synthase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0425|M.bovis_AF2122/97 --------------MAESKLVIGDRSFASRLIMGTGGATNLAVLEQALIA
Rv0417|M.tuberculosis_H37Rv --------------MAESKLVIGDRSFASRLIMGTGGATNLAVLEQALIA
MMAR_0720|M.marinum_M --------------MVESKLTIADRSFASRLIMGTGGASNLAVLQEALVA
MUL_2803|M.ulcerans_Agy99 --------------MVESKLMIADRSFASRLIMGTGGASNLAVLQEALVA
MAV_4744|M.avium_104 --------------MVDSKLTIADRSFSSRLIMGTGGAGNLAVLEEALIA
Mflv_0195|M.gilvum_PYR-GCK MVE----------FGTGK-LSIAGREFGSRLILGTGGAANLAVLEAALVA
Mvan_0710|M.vanbaalenii_PYR-1 MAESEVSDFADSDFATGK-LSIAGREFGSRLILGTGGAANLAVLEAALVA
TH_2314|M.thermoresistible__bu VAS----------PDTDK-LTIAGRTFGSRLILGTGGAANLAILEEALVA
MAB_4216c|M.abscessus_ATCC_199 -------------MGTGESLCIAGRTFGSRLIMGTGGAANLSILERALVA
MSMEG_0793|M.smegmatis_MC2_155 --------------MADSVLRIGGREFGSRLIMGTGGAPNLSVLEEALIA
MLBr_00297|M.leprae_Br4923 --------------MVESKFTIGDRTFTSRLIMGTGGAANLAILEEALIA
. . : *..* * ****:***** **::*: **:*
Mb0425|M.bovis_AF2122/97 SGTELTTVAIRRVDADGGTGLLDLLNRLGITPLPNTAGCRSAAEAVLTAQ
Rv0417|M.tuberculosis_H37Rv SGTELTTVAIRRVDADGGTGLLDLLNRLGITPLPNTAGSRSAAEAVLTAQ
MMAR_0720|M.marinum_M SGTELTTVAIRRVDAEGGTGLLDLLNRLGITPLPNTAGCRSAAEAVLTAQ
MUL_2803|M.ulcerans_Agy99 SGTELTTVAIRRVDAEGGTGLLDLLNRLGITPLPNTAGCRSAAEAVLTAQ
MAV_4744|M.avium_104 SGTELTTVAIRRVDAEGGTGLLDLLNRLGITPLPNTAGCRGAAEAVLTAQ
Mflv_0195|M.gilvum_PYR-GCK SGTELTTVAMRRIDADGGTGVLDLLTRLGITPLPNTAGCRGAAEAVMTAQ
Mvan_0710|M.vanbaalenii_PYR-1 SGTELTTVAMRRVDAEGGTGVLDLLTRLGISPLPNTAGCRGAAEAVMTAQ
TH_2314|M.thermoresistible__bu SGTELTTVAMRRVDAEGGTGVLELLNRLGITPLPNTAGCRGAAEAVYTAQ
MAB_4216c|M.abscessus_ATCC_199 SGTELTTVAIRRVDAAGGTGVLDLLKRLDIVPLPNTAGCRGAAEAVMTAR
MSMEG_0793|M.smegmatis_MC2_155 SGTELTTVAMRRVDAETGTGVLDLLNRLGIAALPNTAGCRGAAEAVLTAQ
MLBr_00297|M.leprae_Br4923 SGTELTTVAIRRVDTDGGSGLLKLLSRLDIMPLPNTAGCRSAAEAVLTAQ
*********:**:*: *:*:*.**.**.* .******.*.***** **:
Mb0425|M.bovis_AF2122/97 LAREALNTNWVKLEVIADERTLWPDAVELVRAAEQLVDDGFVVLPYTTDD
Rv0417|M.tuberculosis_H37Rv LAREALNTNWVKLEVIADERTLWPDAVELVRAAEQLVDDGFVVLPYTTDD
MMAR_0720|M.marinum_M LAREALATNWVKLEVIADERTLLPDAIELVRAAEQLVDDGFVVLPYTNDD
MUL_2803|M.ulcerans_Agy99 LAREALATNWVKLEVIADERTLLPDAIELVRAAEQLVDDGFVVLPYTNDD
MAV_4744|M.avium_104 LAREALNTNWIKLEVIADERTLLPDAVELVRAAEQLVDDGFVVLPYTNDD
Mflv_0195|M.gilvum_PYR-GCK LAREALQTNWVKLEVIADERTLLPDAVELVRAAEQLVDDGFAVLPYTNDD
Mvan_0710|M.vanbaalenii_PYR-1 LAREALQTNWVKLEVIADERTLLPDAIELVRAAEQLVDDGFVVLPYTNDD
TH_2314|M.thermoresistible__bu LGREALDTDWVKLEVIADERTLLPDAVELVRAAEQLVDDGFTVLPYTNDD
MAB_4216c|M.abscessus_ATCC_199 LAREALETDWIKLEVIADERTLLPDAVELVRAAEQLVDDGFVVLPYTTDD
MSMEG_0793|M.smegmatis_MC2_155 LAREALGTDMVKLEVIADERTLLPDAVELVKAAEQLVDDGFTVLPYTNDD
MLBr_00297|M.leprae_Br4923 LAREALNTNWIKLEVIADERTLLPDGLELVRAAEQLVDAGFVVLPYTNDD
*.**** *: :*********** **.:***:******* **.*****.**
Mb0425|M.bovis_AF2122/97 PVLARRLEDTGCAAVMPLGSPIGTGLGIANPHNIEMIVAGARVPVVLDAG
Rv0417|M.tuberculosis_H37Rv PVLARRLEDTGCAAVMPLGSPIGTGLGIANPHNIEMIVAGARVPVVLDAG
MMAR_0720|M.marinum_M PVLARRLEDTGCAAVMPLGSPIGTGLGITNPHNIEMIVASAGVPVVLDAG
MUL_2803|M.ulcerans_Agy99 PVLARRLEDTGCAAVMPLGSPIGTGLGITNPHNIEMIVASAGVPVVLDAG
MAV_4744|M.avium_104 PVLARRLEDAGCAAVMPLGSPIGTGLGITNPHNIEMIVAGAGVPVVLDAG
Mflv_0195|M.gilvum_PYR-GCK PVLARRLEATGCVAVMPLGSPIGTGLGIANPHNIEMIVDSAQVPVILDAG
Mvan_0710|M.vanbaalenii_PYR-1 PVLARRLEATGCAAVMPLGSPIGTGLGIANPHNIEMIVDQASVPVILDAG
TH_2314|M.thermoresistible__bu PVLAKRLEDVGCAAVMPLGSPIGTGLGISNPHNIEMIVAQSNVPVILDAG
MAB_4216c|M.abscessus_ATCC_199 PVLARRLEDVGCAAVMPLGSPIGTGLGITNTHNIEMIVAAAGVPVILDAG
MSMEG_0793|M.smegmatis_MC2_155 PVLARRLEDIGCAAVMPLGSPIGTGLGISNPHNIEMIVAAAGVPVVLDAG
MLBr_00297|M.leprae_Br4923 PALAHRLEGTGCAAVMPLGSPIGTGLGINNPHNIEIIVAQARVPVVLDAG
*.**:*** **.*************** *.****:** : ***:****
Mb0425|M.bovis_AF2122/97 IGTASDAALAMELGCDAVLLASAVTRAADPPAMAAAMAAAVTAGYLARCA
Rv0417|M.tuberculosis_H37Rv IGTASDAALAMELGCDAVLLASAVTRAADPPAMAAAMAAAVTAGYLARCA
MMAR_0720|M.marinum_M IGTASDAALAMELGCDAVLLATAVTRAADPAAMAAAMSAAVTAGYLARRA
MUL_2803|M.ulcerans_Agy99 IGTASDAALAMELGCDALLLATAVTRAADPAAMAAAMSAAVTAGYLARCA
MAV_4744|M.avium_104 IGTASDAALAMELGCDAVLLATAVTRAGDPAAMAAAMSAAVTAGYLARRA
Mflv_0195|M.gilvum_PYR-GCK IGTASDAALAMELGCDAVLLASAVTRAADPAAMAAAMSAAVRAGHLARHA
Mvan_0710|M.vanbaalenii_PYR-1 IGTASDAALAMELGCDAVLLASAVTRSADPPAMAAAMAAAVRAGYLARRA
TH_2314|M.thermoresistible__bu IGTASDAALAMELGCDAVLLATAVTRAGDPPKMAAAMAAAVTAGRLARQA
MAB_4216c|M.abscessus_ATCC_199 IGTASDAALAMELGCDAVLLATAVTRAADPERMAAAMAAAVNAGHLARHA
MSMEG_0793|M.smegmatis_MC2_155 IGTASDAALAMELGCDAVLLATAVTRASDPPTMAAAMASAVTAGHLARQA
MLBr_00297|M.leprae_Br4923 IGTTSDAALAMELGCDAVLLASAVTRAVDPPTMAAAMASAVTAGYLARRA
***:*************:***:****: ** *****::** ** *** *
Mb0425|M.bovis_AF2122/97 GRIPKRFWAQASSPAR------------
Rv0417|M.tuberculosis_H37Rv GRIPKRFWAQASSPAR------------
MMAR_0720|M.marinum_M GRIPKRFWAQASSPTLVTTQSPGAESGN
MUL_2803|M.ulcerans_Agy99 GRIPKRFWAQASSPTLVTTQSPGAESGN
MAV_4744|M.avium_104 GRIPKRFWAQASSPPR------------
Mflv_0195|M.gilvum_PYR-GCK GRIPKRFWAHASSPAFE-----------
Mvan_0710|M.vanbaalenii_PYR-1 GRIPKRFWAQASSPSL------------
TH_2314|M.thermoresistible__bu GRIPKRFWAQASSPPL------------
MAB_4216c|M.abscessus_ATCC_199 GRIPRRFWAQASSPAPDRPLEQ------
MSMEG_0793|M.smegmatis_MC2_155 GRIPKRFWAQASSPAL------------
MLBr_00297|M.leprae_Br4923 GRIPKRFWAQASSPELMRTGEELGN---
****:****:****