For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LELKWVKRGGRIAGLPRRLPPAARKSGIESSARGTDKLGALPLAEAYGQPGAARKPKQVRAASWQGDLYA YLVRWRDPGIVVEFGSAFGVSGMYLSSELRRGWLYSFEINPSWADIAERNIKSVTDRCTLTRGAFEDHVD DIPAPIDMALVDGIHSYDFVARQWQLLKPRMAPGGWVLFDDIDFNPGMGKAWQDICAEPHVVGAVEVSKR LGLIELAP
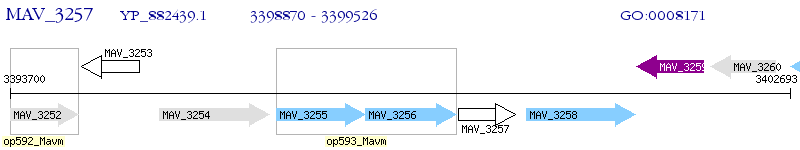
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_3257 | - | - | 100% (218) | hypothetical protein MAV_3257 |
| M. avium 104 | MAV_5200 | - | 4e-09 | 31.40% (121) | O-methyltransferase, family protein 3 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3463 | - | 5e-07 | 29.13% (127) | O-methyltransferase family protein |
| M. marinum M | MMAR_0313 | - | 3e-06 | 29.27% (123) | O-methyltransferase |
| M. smegmatis MC2 155 | MSMEG_4739 | - | 2e-61 | 55.25% (219) | hypothetical protein MSMEG_4739 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_4809 | - | 2e-06 | 29.27% (123) | O-methyltransferase |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0313|M.marinum_M -----MGTTLQDAKVAAALDRMYAAAAN---EMTALRERRAEIERLWSGD
MUL_4809|M.ulcerans_Agy99 -----MGTTLQDAKVAAALDRMYAAAAN---EMTALRERRAEIERLWSGD
MAB_3463|M.abscessus_ATCC_1997 -----MTATLHEPKFAAIIDRLFRNAAN---DPIPT-DR----DTFHKKS
MAV_3257|M.avium_104 ---MELKWVKRGGRIAGLPRRLPPAARKSGIESS-ARGTDKLGALPLAEA
MSMEG_4739|M.smegmatis_MC2_155 MFTRSLKWITRGGWIADMPSRFPPAPHKDRIEAILAANHEQLGPQALAPE
: : .* *: . : :
MMAR_0313|M.marinum_M AQQRADAMSEFYIPVTPEAGRLMYALVRAIRPTTVVEFGMSFGISTIHLA
MUL_4809|M.ulcerans_Agy99 AQQRADAMSEFYIPVTPEAGRLMYALVRAIRPTTVVEFGMSFGISTIHLA
MAB_3463|M.abscessus_ATCC_1997 VEERIELFENIYVPVAPDAGRLLYSLVRAIKPRTIVEYGLSYGISTLHSA
MAV_3257|M.avium_104 YGQPGAARKPKQVRAASWQGDLYAYLVRWRDPGIVVEFGSAFGVSGMYLS
MSMEG_4739|M.smegmatis_MC2_155 YDMPEAKFTPRDVSSPPFQGDLYAWLVMQRKPQTVVEFGSGFGVSGMYFG
: .. * * ** * :**:* .:*:* :: .
MMAR_0313|M.marinum_M AAVRDNGSGRVVTTELNASKIAAAKQTFADTGLADVITVAEGDALATLTT
MUL_4809|M.ulcerans_Agy99 AAVRDNGSGRVVTTELNASKIAAAKQTFADTELADVITVAEGDALATLTT
MAB_3463|M.abscessus_ATCC_1997 AAVRDNGFGRIITTEMNTAKIAASRATFAEAGVSDLITILEGDARETLKT
MAV_3257|M.avium_104 SELRR---GWLYSFEINPSWADIAERNIKS--VTDRCTLTRGAFEDHVDD
MSMEG_4739|M.smegmatis_MC2_155 AGIESNGTGHLYSFEINDSWAAVAERSIAQ--LTQRFTLTRGAFEEHVTE
: :. * : : *:* : :. .: . ::: *: .* :
MMAR_0313|M.marinum_M VPGPVQFVLLDGWK--DLYLPVVRLLEPVLPAGALILADNTESAG--SRP
MUL_4809|M.ulcerans_Agy99 VPGPVQFVLLDGWK--DLYLPVVRLLEPVLPAGALILADNTESAG--SRP
MAB_3463|M.abscessus_ATCC_1997 IDSPIDFLLLDGWP--DLDLTILNIVEDKLAPGALVIADNVGFES--SKP
MAV_3257|M.avium_104 IPAPIDMALVDGIHSYDFVARQWQLLKPRMAPGGWVLFDDIDFNPGMGKA
MSMEG_4739|M.smegmatis_MC2_155 VPGPIDIAFVDGIHTYDFVMQQWEILKPLMSPGGLVLFDDITYGQGMHEA
: .*::: ::** *: .::: :..*. :: *: ..
MMAR_0313|M.marinum_M YLDYIRDPANGYVSFNFPARSSDSMEISCRTAD
MUL_4809|M.ulcerans_Agy99 YLDYIRDPANGYVSFNFPARSSDSMEISCRTAD
MAB_3463|M.abscessus_ATCC_1997 YLEYVRNPENGYVSVASPIGECMELSTSCKK--
MAV_3257|M.avium_104 WQDICAEPH---VVGAVEVSK--RLGLIELAP-
MSMEG_4739|M.smegmatis_MC2_155 WLEIGRSDD---VVGAVEFRRNNRLGLAECR--
: : . * :