For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MLRDTVRRFMDAHVHPVEEKLDHDTVGLPREQLVALQAKAREAGLWSLQTPEEYGGAGLSVLGQVVVAEE AAKCRMGAFFPALGAFGGNPPNIMFKATPEQFEKYARPVIDGIMSKAYTAITEASGGSDPARAIKLKATR DGSSYLLNGSKMWISHAPDADWGVVYARTGEGRHGISAFILEKGTPGVTFSRIPVMASFAPYELHFDNVR LPAEQLIGEEGQGFSLAGEFLVHGRIVYAAGPIGIAQSALDMACRWARDRDVFGGRLADKQGIQWMLVDS EVDLRAARLLMYQAASNADLGRDVRVDASVAKMFGTEAAYRVLDRCIQIHGALGISTELPLERWFRDLRV KRLGEGATEVQREIIARSLVR*
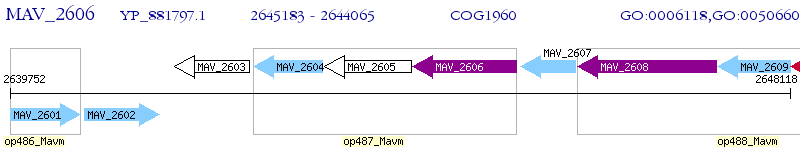
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_2606 | - | - | 100% (372) | acyl-CoA dehydrogenase |
| M. avium 104 | MAV_1896 | - | 0.0 | 90.32% (372) | acyl-CoA dehydrogenase |
| M. avium 104 | MAV_4243 | - | 9e-51 | 32.01% (378) | acyl-CoA dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3302c | fadE25 | 5e-52 | 32.54% (378) | acyl-CoA dehydrogenase FADE25 |
| M. gilvum PYR-GCK | Mflv_4943 | - | 1e-48 | 32.72% (379) | acyl-CoA dehydrogenase domain-containing protein |
| M. tuberculosis H37Rv | Rv3274c | fadE25 | 6e-52 | 32.54% (378) | acyl-CoA dehydrogenase FADE25 |
| M. leprae Br4923 | MLBr_00737 | fadE25 | 3e-51 | 33.07% (378) | putative acyl-CoA dehydrogenase |
| M. abscessus ATCC 19977 | MAB_0328 | - | 4e-54 | 37.33% (375) | putative acyl-CoA dehydrogenase |
| M. marinum M | MMAR_1262 | fadE25 | 2e-49 | 31.75% (378) | acyl-CoA dehydrogenase FadE25 |
| M. smegmatis MC2 155 | MSMEG_0108 | - | 3e-53 | 35.78% (341) | acyl-CoA dehydrogenase |
| M. thermoresistible (build 8) | TH_3846 | - | 6e-54 | 32.63% (380) | PUTATIVE acyl-CoA dehydrogenase |
| M. ulcerans Agy99 | MUL_2626 | fadE25 | 2e-50 | 32.28% (378) | acyl-CoA dehydrogenase FadE25 |
| M. vanbaalenii PYR-1 | Mvan_1476 | - | 1e-49 | 33.25% (379) | acyl-CoA dehydrogenase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3302c|M.bovis_AF2122/97 MVGWAGNPSFDLFKLPEEHDEMRSAIRALAEKEIAPHAAEVDEKARFPEE
Rv3274c|M.tuberculosis_H37Rv MVGWAGNPSFDLFKLPEEHDEMRSAIRALAEKEIAPHAAEVDEKARFPEE
MLBr_00737|M.leprae_Br4923 MVGWSGNPLFDLFKLPEEHNELRATIRALAEKEIAPHAADVDQRARFPEE
MMAR_1262|M.marinum_M MAGWAGNPSFDLFQLPEEHQELRAAIRALAEKEIAPHAADVDENSRFPQE
MUL_2626|M.ulcerans_Agy99 MAGWAGNPSFDLFQLPEEHQELRAAIRALAEKEIAPHAADVDENSRFPQE
TH_3846|M.thermoresistible__bu MAAWSGNPSFELFQLPEEHIALREAIRALAEKEIAPYAAEVDEKARFPEE
Mflv_4943|M.gilvum_PYR-GCK -------MPVDRLLPTDDARDLVELARQIADKVLDPIVDRHEKDETYPEG
Mvan_1476|M.vanbaalenii_PYR-1 -------MAVDRLLPTDEARELVDLARQIADKALDPIVDQHEKDETYPDG
MSMEG_0108|M.smegmatis_MC2_155 -------MSVDRLLPSEEARDLIALTREVADKVLDPIVDEHERNETYPEG
MAV_2606|M.avium_104 --------------MMLRDTVRRFMDAHVHPVEEKLDHDTVGLPREQLVA
MAB_0328|M.abscessus_ATCC_1997 ----MIDFSIPEELAAKAQRVREFVTETVIPYERDPRLTAHGPTEELRDE
:
Mb3302c|M.bovis_AF2122/97 ALVALNSSGFNAVHIPEEYGGQGADSVATCIVIEEVARVDASASLIPAVN
Rv3274c|M.tuberculosis_H37Rv ALVALNSSGFNAVHIPEEYGGQGADSVATCIVIEEVARVDASASLIPAVN
MLBr_00737|M.leprae_Br4923 ALAALNASGFNAIHVPEEYGGQGADSVAACIVIEEVARVDASASLIPAVN
MMAR_1262|M.marinum_M ALDALNASGFNAVHVPEEYGGQGADSVAACIVIEEVARVDASASLIPAVN
MUL_2626|M.ulcerans_Agy99 ALDALNASGFNALHVPEEYGGQGADSVAACIVIEEVARVDASASLIPAVN
TH_3846|M.thermoresistible__bu ALAALNSSGFSAIHVPEEYGGQGADSVATCIVIEEVARVDCSASLIPAVN
Mflv_4943|M.gilvum_PYR-GCK VFATLGEAGLLSLPYPEEWGGGGQPYEVYLQVLEELAARWAAVAVAVSVH
Mvan_1476|M.vanbaalenii_PYR-1 VFATLGEAGLLSLPYPEEWGGGGQPYEVYLQVLEELAARWAAVAVAVSVH
MSMEG_0108|M.smegmatis_MC2_155 VFAQLGAAGLLSLPQPEQWGGGGQPYEVYLQVLEEIAARWAAVAVAVSVH
MAV_2606|M.avium_104 LQAKAREAGLWSLQTPEEYGGAGLSVLGQVVVAEEAAKCRMGAFFPALGA
MAB_0328|M.abscessus_ATCC_1997 LVGKARAAGLLTVQAPESYGGWGLSHIGQAVIFEAAGWSTLGPIAMNCAA
:*: :: **.:** * : * . .
Mb3302c|M.bovis_AF2122/97 KLGTMG--LILRGSEELKKQVLPALAAEGAMASYALSERE-AGSDAA-SM
Rv3274c|M.tuberculosis_H37Rv KLGTMG--LILRGSEELKKQVLPALAAEGAMASYALSERE-AGSDAA-SM
MLBr_00737|M.leprae_Br4923 KLGTMG--LILRGSEELKKQVLPSLAAEGAMASYALSERE-AGSDAA-SM
MMAR_1262|M.marinum_M KLGTMG--LILRGSDDLKKQVLPSLADGAAMASYALSERE-AGSDAG-AM
MUL_2626|M.ulcerans_Agy99 KLGTMG--LILRGSDELKKQVLPSLADGAAMASYALSERE-AGSDAG-AM
TH_3846|M.thermoresistible__bu KLGTMG--LILRGSEELKKQVLPAVASGEAMASYALSERE-AGSDAA-SM
Mflv_4943|M.gilvum_PYR-GCK SLSCHP--LMSFGSDEQRQRWLPDMLGGSTIGAYSLSEPQ-AGSDAA-AL
Mvan_1476|M.vanbaalenii_PYR-1 SLSCHP--LMSFGTDEQRQRWLPEMLGGSTIGAYSLSEPQ-AGSDAA-AL
MSMEG_0108|M.smegmatis_MC2_155 SLSSHP--LLAFGTDEQKQRWLPGMLSGEQIGAYSLSEPQ-AGSDAA-AL
MAV_2606|M.avium_104 FGGNPPNIMFKATP-EQFEKYARPVIDGIMSKAYTAITEASGGSDPARAI
MAB_0328|M.abscessus_ATCC_1997 PDEGNMFLLGKITNEEQSEKFLRPVIEGYQRSAFAMTEPDGAGSDPS-QL
: : :: : ::: .***.. :
Mb3302c|M.bovis_AF2122/97 RTRAKADGDHWILNGAKCWITNGGKSTWYTVMAVTDPDRGAN-GISAFMV
Rv3274c|M.tuberculosis_H37Rv RTRAKADGDHWILNGAKCWITNGGKSTWYTVMAVTDPDRGAN-GISAFMV
MLBr_00737|M.leprae_Br4923 RTRAKADGDDWILNGFKCWITNGGKSTWYTVMAVTDPDKGAN-GISAFIV
MMAR_1262|M.marinum_M RTRAKADGDDWILNGTKCWITNGGKSSWYTVMAVTDPEKGAN-GISAFMV
MUL_2626|M.ulcerans_Agy99 RTRAKADGDDWILNGTKCWITNGGKSSWYTVMAVTDPENGAN-GISAFVV
TH_3846|M.thermoresistible__bu RTRAVADGDDWILNGSKCWITNGGKSTWYTVMAVTDPDKGAN-GISAFMV
Mflv_4943|M.gilvum_PYR-GCK TCRATAVDGGYRITGSKAWITHGGIADFYNLFARTG--EGSQ-GISCFLV
Mvan_1476|M.vanbaalenii_PYR-1 TCKAVAADGGYRINGAKAWITHGGIADFYNLFARTG--EGSQ-GISCFLV
MSMEG_0108|M.smegmatis_MC2_155 RCAARRDGDGFVLNGSKAWITHGGRADFYTLFARTG--EGSK-GVSCFLV
MAV_2606|M.avium_104 KLKATRDGSSYLLNGSKMWISHAPDADWGVVYARTGEGRH---GISAFIL
MAB_0328|M.abscessus_ATCC_1997 KTSATFDGQNFVINGRKWLITGARGAKTWIIMANLEANDHLPEGPTLFLC
* . : :.* * *: . : : * * : *:
Mb3302c|M.bovis_AF2122/97 HKDDEGFTVGPKERKLGIKGSP-TTELYFENCRIPGDRIIGEPGTGFKTA
Rv3274c|M.tuberculosis_H37Rv HKDDEGFTVGPKERKLGIKGSP-TTELYFENCRIPGDRIIGEPGTGFKTA
MLBr_00737|M.leprae_Br4923 HKDDEGFSIGPKEKKLGIKGSP-TTELYFDKCRIPGDRIIGEPGTGFKTA
MMAR_1262|M.marinum_M HKDDEGFVVGPKERKLGIKGSP-TTELYFENCRVPGDRMIGEPGTGFKTA
MUL_2626|M.ulcerans_Agy99 HKDDEGFVVGPKERKLGIKGSP-TTELYFENCRVPGDRMIGEPGTGFKTA
TH_3846|M.thermoresistible__bu HKDDEGFTVGPKERKLGIKGSP-TTELYFENCRIPGDRIIGEPGTGFKTA
Mflv_4943|M.gilvum_PYR-GCK PRDTEGLTFGKPEEKMGLHAVP-TTAAHYDDANLDSDRRIGTEGQGLQIA
Mvan_1476|M.vanbaalenii_PYR-1 PRDTEGLTFGKPEEKMGLHAVP-TTAAHYENAFVEADRRIGAEGQGLQIA
MSMEG_0108|M.smegmatis_MC2_155 PGDLPGLSFGKPEEKMGLHAVP-TTSAFYDNARIDADRLIGTEGQGLSIA
MAV_2606|M.avium_104 EKGTPGVTFSRIP-VMAS-FAP--YELHFDNVRLPAEQLIGEEGQGFSLA
MAB_0328|M.abscessus_ATCC_1997 DGETEGIVIERIMNTMDRNYAEGHAVVRFDNLTLPREALLGETGQAFRYA
*. . : ::. : : :* * .: *
Mb3302c|M.bovis_AF2122/97 LATLDHTRPTIGAQAVGIAQGALDAAIAYTKDRKQFGESISTFQAVQFML
Rv3274c|M.tuberculosis_H37Rv LATLDHTRPTIGAQAVGIAQGALDAAIAYTKDRKQFGESISTFQAVQFML
MLBr_00737|M.leprae_Br4923 LATLDHTRPTIGAQAVGIAQGALDAAIVYTKDRKQFGESISTFQSIQFML
MMAR_1262|M.marinum_M LATLDHTRPTIGAQAVGIAQGALDAAIAYTKDRKQFGKSISDFQAVQFML
MUL_2626|M.ulcerans_Agy99 LATLDHTRPTIGAQAVGIAQGALDAAIAYTKDRKQFGKSISDFQAVQFML
TH_3846|M.thermoresistible__bu LATLDHTRPTIGAQAVGIAQGALDAAIAYTKERKQFGRPVSDNQGVQFML
Mflv_4943|M.gilvum_PYR-GCK FSALDSGRLGIAAVAVGLAQAALDEAVAYSQERTTFGRKIIDHQGLAFLL
Mvan_1476|M.vanbaalenii_PYR-1 FSALDSGRLGIAAVAVGLAQAALDAAVDYSQERTTFGRKIIDHQGLAFLL
MSMEG_0108|M.smegmatis_MC2_155 FSALDSGRLGIAAVAVGIAQAALDEAVRYAHERTTFGRKIIDHQGLGFLL
MAV_2606|M.avium_104 GEFLVHGRIVYAAGPIGIAQSALDMACRWARDRDVFGGRLADKQGIQWML
MAB_0328|M.abscessus_ATCC_1997 QLRLAPARLTHCMRWLGAASRAQDIAVDYARTRTSFGKTIGEHQGVSFML
* * :* *. * * * ::: * ** : *.: ::*
Mb3302c|M.bovis_AF2122/97 ADMAMKVEAARLMVYSAAARAERGEPDLGFISAASKCFASDVAMEVTTDA
Rv3274c|M.tuberculosis_H37Rv ADMAMKVEAARLMVYSAAARAERGEPDLGFISAASKCFASDVAMEVTTDA
MLBr_00737|M.leprae_Br4923 ADMAMKVEAARLIVYAAAARAERGEPDLGFISAASKCFASDIAMEVTTDA
MMAR_1262|M.marinum_M ADMAMKVESARLMVYSAAARAERGESNLGFISAASKCLASDVAMEVTTDA
MUL_2626|M.ulcerans_Agy99 ADMAMKVESARLMVYSAAARAERGESNLGFISAASKCLASDVAMEVTTDA
TH_3846|M.thermoresistible__bu ADMAMKIEAARLMVYSAAARAERGEGDLGFISAASKCFASDVAMEVTTDA
Mflv_4943|M.gilvum_PYR-GCK ADMAAAVDSARATYLDAARRRDAGLP-YSRHASVAKLVATDAAMKVTTDA
Mvan_1476|M.vanbaalenii_PYR-1 ADMAAAVDAARATYLDAARRRDAGLP-YSRHASVAKLTATDAAMKVTTDA
MSMEG_0108|M.smegmatis_MC2_155 ADMAAAVVSARATYLDAARRRDAGLP-YSSQASVAKLVATDAAMKVTTDA
MAV_2606|M.avium_104 VDSEVDLRAARLLMYQAASNADLGRD-VRVDASVAKMFGTEAAYRVLDRC
MAB_0328|M.abscessus_ATCC_1997 ADNEIALHQCRLTIWHACWMMDNGAK-GRHESSMAKSFVSEELFKVTDRC
.* : .* *. : * :: :* :: .* .
Mb3302c|M.bovis_AF2122/97 VQLFGGAGYTTDFPVERFMRDAKITQIYEGTNQIQRVVMSRALLR----
Rv3274c|M.tuberculosis_H37Rv VQLFGGAGYTTDFPVERFMRDAKITQIYEGTNQIQRVVMSRALLR----
MLBr_00737|M.leprae_Br4923 VQLFGGAGYTSDFPVERFMRDAKITQIYEGTNQIQRVVMSRALLR----
MMAR_1262|M.marinum_M VQLFGGAGYTVDFPVERMMRDAKITQIYEGTNQIQRVVMSRALLR----
MUL_2626|M.ulcerans_Agy99 VQLFGGAGYTVDFPVERMMRDAKITQIYEGTNQIQRVVMSRALLR----
TH_3846|M.thermoresistible__bu VQLFGGYGYTQDFPVERMMRDAKITQIYEGTNQIQRVVMSRALLR----
Mflv_4943|M.gilvum_PYR-GCK VQVLGGAGYTRDYRVERYMREAKITQIFEGTNQIQRLVISRHLTRG---
Mvan_1476|M.vanbaalenii_PYR-1 VQVFGGAGYTRDYRVERYMREAKITQIFEGTNQIQRLVISRHLARD---
MSMEG_0108|M.smegmatis_MC2_155 VQVLGGVGYTRDFRVERYMREAKITQIFEGTNQIQRLVISRGLGT----
MAV_2606|M.avium_104 IQIHGALGISTELPLERWFRDLRVKRLGEGATEVQREIIARSLVR----
MAB_0328|M.abscessus_ATCC_1997 VQVLGGIGISDETPVEMIFRDMRAFRLYDGPTEVHKYAIGRQVLRTPRA
:*: *. * : : :* :*: : :: :*..:::: :.* :