For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LKIRDKVFVVTGGANGMGRQVALLLVAKGARVAVVDVDVAGLKETAALTAPQAGRISTHVVDVAERSATS ALPDAVVAAHGAVDGLVNVAGIIHRFARLNDLEFGEIERVLNINLYGVLNVTKAFLPHLLKRPTAHITIV SSMASYAPVPGQAIYGASKAAVKMLAEGLNSELRDTNVRVSVVFPGAVKTDIAVNSGAMTPDENEATARS MRMLPSTVAAAKIVRGIEKDSRRVFVGLDALLLDKLNRLTPASAAKLIWSQMRDVLPPAGQSLR
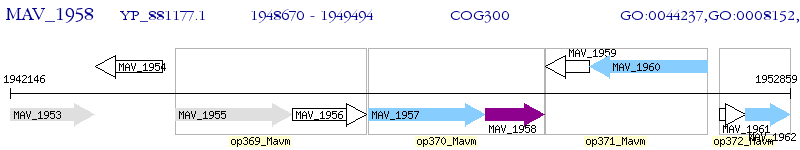
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_1958 | - | - | 100% (274) | short chain dehydrogenase |
| M. avium 104 | MAV_1384 | - | 2e-36 | 38.75% (271) | short chain alcohol dehydrogenase |
| M. avium 104 | MAV_1382 | - | 4e-35 | 36.68% (259) | short chain alcohol dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3112 | - | 5e-41 | 36.94% (268) | short-chain type dehydrogenase/reductase |
| M. gilvum PYR-GCK | Mflv_2216 | - | 2e-34 | 37.55% (277) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv3085 | - | 5e-41 | 36.94% (268) | short-chain type dehydrogenase/reductase |
| M. leprae Br4923 | MLBr_01094 | - | 1e-32 | 34.96% (266) | short chain alcohol dehydrogenase |
| M. abscessus ATCC 19977 | MAB_1388c | - | 4e-37 | 35.66% (272) | short-chain dehydrogenase/reductase |
| M. marinum M | MMAR_4195 | - | 2e-36 | 38.69% (274) | short-chain type dehydrogenase/reductase |
| M. smegmatis MC2 155 | MSMEG_2872 | - | 2e-64 | 48.87% (266) | short-chain dehydrogenase/reductase SDR |
| M. thermoresistible (build 8) | TH_2758 | - | 1e-60 | 48.50% (266) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_4501 | - | 1e-35 | 37.96% (274) | short-chain type dehydrogenase/reductase |
| M. vanbaalenii PYR-1 | Mvan_4479 | - | 1e-36 | 37.73% (273) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4195|M.marinum_M MEGFAGKVAVVTGAGSGIGRALAIELARSGAKLAISDVDTEGLAQTEKLV
MUL_4501|M.ulcerans_Agy99 MEGFAGKVAVVTGAGSGIGRALAIELARSGAKLAISDVDTEGLAQTEKLV
MLBr_01094|M.leprae_Br4923 MEGFAGKVAVVTGAGSGIGQALAIELARSGAKLAISDVDGEGLAQTEGQL
Mflv_2216|M.gilvum_PYR-GCK MQGFAGKVAVVTGAGSGIGQALAIELGRSGALVAISDVDTEGLAVTEERL
Mvan_4479|M.vanbaalenii_PYR-1 MEGFAGKVAVVTGAGSGIGQALAIELGRSGARVAISDVDTEGLAVTEERL
MAB_1388c|M.abscessus_ATCC_199 MEGFSGKVAVVTGAGSGIGRALAVELARSGAKIAISDIDNEGLAETERQV
Mb3112|M.bovis_AF2122/97 MSSFEGKVAVITGAGSGIGRALALNLSEKRAKLALSDVDTDGLAKTVRLA
Rv3085|M.tuberculosis_H37Rv MSSFEGKVAVITGAGSGIGRALALNLSEKRAKLALSDVDTDGLAKTVRLA
MSMEG_2872|M.smegmatis_MC2_155 -MRIADRVFVVTGAGNGMGREVALGLARRGAHVAAVDLDEAGLAGTAERA
TH_2758|M.thermoresistible__bu -VRITDRVFVVTGSGNGMGREVAVELGRRGAHVAAVDIDREGLAGTAELV
MAV_1958|M.avium_104 -MKIRDKVFVVTGGANGMGRQVALLLVAKGARVAVVDVDVAGLKETAALT
: .:* *:**...*:*: :*: * * :* *:* ** *
MMAR_4195|M.marinum_M TALGAEVKTDRLDVTEREAFLAYADAVNEHFGKVNQIYNNAGIGHTGDVE
MUL_4501|M.ulcerans_Agy99 TALGAEVKTDRLDVTERETFLAYADAVNEHFGKVNQIYNNAGIGHTGDVE
MLBr_01094|M.leprae_Br4923 KAIGASARTDRLDVTEREAFLTYADVVHENFGKVNQIYNNAGIAFTGDVE
Mflv_2216|M.gilvum_PYR-GCK KAIGTHVKSDRLNVTERERFLQYADDVAEHFGKVHQIYNNAGIAFTGDIE
Mvan_4479|M.vanbaalenii_PYR-1 KAIGAQVKSDRLDVTERETFLLYADDVAEHFGSVNQIYNNAGIAFTGDIE
MAB_1388c|M.abscessus_ATCC_199 KALGAEVRADRLNVAEREAFLLYADAIKDHFGKVNQIYNNAGIAYQGEVE
Mb3112|M.bovis_AF2122/97 QALGAQVKSDRLDVAEREAVLAHADAVVAHFGTVHQVYNNAGIAYNGNVD
Rv3085|M.tuberculosis_H37Rv QALGAQVKSDRLDVAEREAVLAHADAVVAHFGTVHQVYNNAGIAYNGNVD
MSMEG_2872|M.smegmatis_MC2_155 RGTGARMSTHVLNVTDRDAVAGLPDAVLDVHGQIDGLVNIAGIAQRFALF
TH_2758|M.thermoresistible__bu RATGARITTHVVDVTDDNAVAALPAAVLDAHGQVDGLVNIAGIAQRFALF
MAV_1958|M.avium_104 APQAGRISTHVVDVAERSATSALPDAVVAAHGAVDGLVNVAGIIHRFARL
. :. ::*:: . . : .* :. : * ***
MMAR_4195|M.marinum_M -VCAFKDIDRVMDVDFGGVLNGTKAFLPYLIASGDGHVINVSSVFGLFSV
MUL_4501|M.ulcerans_Agy99 -VCAFKDIDRVMDVDFGGVLNGTKTFLPYLIASGDGHVINVSSVFGLFSV
MLBr_01094|M.leprae_Br4923 -VSHFKDIERVMDVDYWGVVNGTKAFLPYLISSGDGHVINISSVFGLFSV
Mflv_2216|M.gilvum_PYR-GCK -VSQFKDIERVMDVDFWGVVNGTKAFLPHLIASGDGHVVNVSSVFGLFSV
Mvan_4479|M.vanbaalenii_PYR-1 -ITQFKDIERVMDVDYWGVVNGTKAFLPHLIASGDGHVVNVSSVFGLFSV
MAB_1388c|M.abscessus_ATCC_199 -ESQFKDIERIIDVDFWGVVNGTKAFLPHLIASGDGHVVNVSSLFGVLSM
Mb3112|M.bovis_AF2122/97 -KSEFKDIERIIDVDFWGVVNGTKAFLPHVIASGDGHIVNISSLFGLIAV
Rv3085|M.tuberculosis_H37Rv -KSEFKDIERIIDVDFWGVVNGTKAFLPHVIASGDGHIVNISSLFGLIAV
MSMEG_2872|M.smegmatis_MC2_155 TDLEADQIDRVTTVNFLGTVQMCRAFLPVLRQRPEANITNMSSLSALVPF
TH_2758|M.thermoresistible__bu SDLGAEALDRVPVVNFTGTVRMCRAFLPILLARPEANITNMSSLSALVPF
MAV_1958|M.avium_104 NDLEFGEIERVLNINLYGVLNVTKAFLPHLLKRPTAHITIVSSMASYAPV
::*: :: *.:. ::*** : .:: :**: . ..
MMAR_4195|M.marinum_M SGQAAYNAAKFAVRGFTEALRQEMILAGHPVGVTTVHPGGIKTAIARNAT
MUL_4501|M.ulcerans_Agy99 SGQAAYNAAKFAVRGFTEALRQEMILAGHPVGVTTVHPGGIKTAIARNAT
MLBr_01094|M.leprae_Br4923 PGQAAYNSAKFAVRGFTEALREEMALAGRPVNVTTVYPGGIKTAIARNAT
Mflv_2216|M.gilvum_PYR-GCK PGQGAYNAAKFAVRGFTEALRQEMALAGHPVKVSCVHPGGIKTAIARNAG
Mvan_4479|M.vanbaalenii_PYR-1 PGQAAYNSAKFAVRGFTEALRQEMALAGHPVKVSCVHPGGIKTAIARNAS
MAB_1388c|M.abscessus_ATCC_199 PGQSAYNSAKFAVRGFTESLRQEMIIGKKPVAVTCVHPGGIKTAIARNST
Mb3112|M.bovis_AF2122/97 PGQSAYNAAKFAVRGFTEALRQEMLVARHPVKVTCVHPGGIKTAVARNAT
Rv3085|M.tuberculosis_H37Rv PGQSAYNAAKFAVRGFTEALRQEMLVARHPVKVTCVHPGGIKTAVARNAT
MSMEG_2872|M.smegmatis_MC2_155 ASQVLYSASKAAVQQFSEGLDAELADTN--VRVVTVFPGQVSTNLAQNS-
TH_2758|M.thermoresistible__bu ASQLLYSASKAAVKQFSEGLDAELVGTD--VRVVTVFPGNVATELAKNS-
MAV_1958|M.avium_104 PGQAIYGASKAAVKMLAEGLNSELRDTN--VRVSVVFPGAVKTDIAVNS-
..* *.::* **: ::*.* *: * * *.** : * :* *:
MMAR_4195|M.marinum_M AAEGLDSDELAKMFDKRVARTSPERAAKIILGAVRKNKARVLVGPDAKAL
MUL_4501|M.ulcerans_Agy99 AAEGLDSDELAKMFDKRVARTSPERAAKIILGAVRKNKARVLVGPDAKAL
MLBr_01094|M.leprae_Br4923 AAEGLDVSKIASRFDTWVAHTSPQHAARIILKAVRKKKARVLVGPDAKVA
Mflv_2216|M.gilvum_PYR-GCK AAEGLDAEELAKTFDKKLASTTPEKAAQVILDGVRKNKARILIGNDAKMF
Mvan_4479|M.vanbaalenii_PYR-1 AAEGLDAEELAKAFDKKLASTTPEKAAKIILDGVRKNRARILVGNDAKVF
MAB_1388c|M.abscessus_ATCC_199 TAEGYDQQAMAAMFDKYLANTSPEAAARIILTAVRKKKPRVLVGPDAKIL
Mb3112|M.bovis_AF2122/97 VADGEDQQTFAEFFDRRLALHSPEMAAKTIVNGVAKGQARVVVGLEAKAV
Rv3085|M.tuberculosis_H37Rv VADGEDQQTFAEFFDRRLALHSPEMAAKTIVNGVAKGQARVVVGLEAKAV
MSMEG_2872|M.smegmatis_MC2_155 ---GVKMLD---AGGRRAPVTTPEAAGRKIVTGIAKDRYRVIIGTDAHVL
TH_2758|M.thermoresistible__bu ---GVEMLD---AGSRRVYATSPEAAGRKIVAGIARDRYRVIIGADAHVL
MAV_1958|M.avium_104 ---GAMTPDENEATARSMRMLPSTVAAAKIVRGIEKDSRRVFVGLDALLL
* .. *. *: .: : *:.:* :*
MMAR_4195|M.marinum_M DIVVRLTG-SGYQRLFMPVLGRLVPASHR---
MUL_4501|M.ulcerans_Agy99 DIVVRLTG-SGYQRLFMPVLGRLVPASHR---
MLBr_01094|M.leprae_Br4923 NVVVRFSGGAGYQRLFAQVASRLILNQR----
Mflv_2216|M.gilvum_PYR-GCK EIAIRALG-AGYQSVFPKVVARLTPPAK----
Mvan_4479|M.vanbaalenii_PYR-1 DVVVRVLG-AGYQSLFPKAVARLTPPAK----
MAB_1388c|M.abscessus_ATCC_199 DVIVRLTG-ARYQDIFSVVTRFILPRPGKK--
Mb3112|M.bovis_AF2122/97 DVLARIMG-SSYQRLVAAGVAKFFPWAK----
Rv3085|M.tuberculosis_H37Rv DVLARIMG-SSYQRLVAAGVAKFFPWAK----
MSMEG_2872|M.smegmatis_MC2_155 YTLSRLAP-RRTARMVAKQIKSVL--------
TH_2758|M.thermoresistible__bu HALARIAP-RRTARLVARQITSVLREQ-----
MAV_1958|M.avium_104 DKLNRLTP-ASAAKLIWSQMRDVLPPAGQSLR
* :. .