For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTASRSPATDLPKSWDPAAAEYAIYRQWVDAGYFTANPASDKPGYSIVLPPPNVTGSLHMGHALEHTMMD ALTRRKRMQGYEVLWQPGMDHAGIATQSVVEKQLAVDGKTKEDFGRELFIEKVWDWKRESGGAIGGQMRR LGDGVDWSRDRFTMDEGLSRAVRTIFKRLYDAGLIYRAERLVNWSPVLQTALSDIEVNYEEVEGELVSFR YGSLDDSGPHIVVATTRVETMLGDTAIAVHPDDERYRHLVGSSLPHPFVDRQLLIVADEHVDPEFGTGAV KVTPAHDPNDFEIGLRHQLPMISIMDTKGRIADTGTQFDGMDRFAARVAVREALAAQGRIVEEKRPYLHS VGHSERSGEPIEPRLSLQWWVRVESLAKAAGDAVRNGDTVIHPTSMEPRWFAWVDDMHDWCISRQLWWGH RIPIWYGPNGEQRCVGPDETPPEGWEQDPDVLDTWFSSALWPFSTLGWPEKTPELEKFYPTSVLVTGYDI LFFWVARMMMFGTFVGDDDAITLDGRRGPQVPFTDVFLHGLIRDESGRKMSKSKGNVIDPLDWVDMFGAD ALRFTLARGASPGGDLAIGEDHVRASRNFCTKLFNATRYALLNGAQLAQLPPLDELTDADRWILGRLEEV RAEVDSAFDNYEFSRACESLYHFAWDEFCDWYVELAKTQLAEGITHTTAVLATTLDTLLRLLHPVIPFIT EALWQALTGNESLVIADWPRSSGVGLDQVATQRITDMQKLVTEVRRFRSDQGLADRQKVPARLAGVTESD LDTQVSAVTSLAWLTDAGPDFRPSASVEVRLRGGTVVVELDTSGSIDVAAERRRLEKDLAAAHKELASTT AKLANEDFLAKAPPHVVDKIRDRQRLAQEESERINARLAVLQ
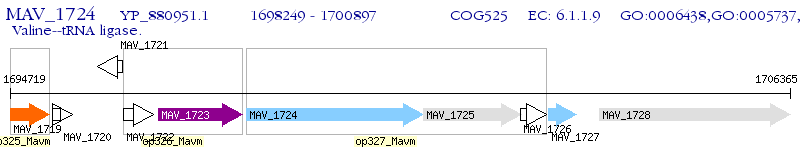
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_1724 | valS | - | 100% (882) | valyl-tRNA synthetase |
| M. avium 104 | MAV_3235 | ileS | 3e-58 | 24.53% (962) | isoleucyl-tRNA synthetase |
| M. avium 104 | MAV_1145 | metG | 2e-11 | 27.70% (213) | methionyl-tRNA synthetase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2475c | valS | 0.0 | 88.34% (875) | valyl-tRNA synthetase |
| M. gilvum PYR-GCK | Mflv_2630 | valS | 0.0 | 80.84% (877) | valyl-tRNA synthetase |
| M. tuberculosis H37Rv | Rv2448c | valS | 0.0 | 88.34% (875) | valyl-tRNA synthetase |
| M. leprae Br4923 | MLBr_01472 | valS | 0.0 | 84.76% (886) | valyl-tRNA synthetase |
| M. abscessus ATCC 19977 | MAB_1603 | valS | 0.0 | 80.11% (880) | valyl-tRNA synthetase |
| M. marinum M | MMAR_3772 | valS | 0.0 | 88.04% (886) | valyl-tRNA synthetase protein ValS |
| M. smegmatis MC2 155 | MSMEG_4630 | valS | 0.0 | 83.55% (833) | valyl-tRNA synthetase |
| M. thermoresistible (build 8) | TH_0609 | valS | 0.0 | 81.54% (872) | PROBABLE VALYL-tRNA SYNTHASE PROTEIN VALS (VALYL-tRNA |
| M. ulcerans Agy99 | MUL_3719 | valS | 0.0 | 87.70% (886) | valyl-tRNA synthetase |
| M. vanbaalenii PYR-1 | Mvan_3951 | valS | 0.0 | 82.45% (883) | valyl-tRNA synthetase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2630|M.gilvum_PYR-GCK MTSQPHANGDARSVPELDALPKSWDPGTVESDLYDGWVEAGYFTADPSSD
Mvan_3951|M.vanbaalenii_PYR-1 MTSQPTANGDAPRAPELDALPKSWDPGAVESDLYDGWVKAGYFTADPTSA
MSMEG_4630|M.smegmatis_MC2_155 MTASPENR--------ADALPKSWDPGAVEAELYQGWVDAGYFTADPASD
TH_0609|M.thermoresistible__bu ----------------VESLPKTWDPAAVETELYEGWVRAGYFTADPSSS
MAB_1603|M.abscessus_ATCC_1997 MTQTPDTN-------AASSLPTSWDPGAVEAELYQGWVDAGYFTADPSSD
Mb2475c|M.bovis_AF2122/97 ------------------MLPKSWDPAAMESAIYQKWLDAGYFTADPTST
Rv2448c|M.tuberculosis_H37Rv ------------------MLPKSWDPAAMESAIYQKWLDAGYFTADPTST
MMAR_3772|M.marinum_M MTASADSA--------ADQLPKSWDPAAMEGDIYQKWADAGYFVADPTSS
MUL_3719|M.ulcerans_Agy99 MTASADSA--------ADQLPKSWDPAAMEGDIYQKWADAGYFVADPTSS
MLBr_01472|M.leprae_Br4923 MTTSPSSS--------ADLLPKSWDPGAMESVIYQKWLNSGYFAADPAST
MAV_1724|M.avium_104 MTAS-RSP--------ATDLPKSWDPAAAEYAIYRQWVDAGYFTANPASD
**.:***.: * :* * :***.*:*:*
Mflv_2630|M.gilvum_PYR-GCK KPAYSIVLPPPNVTGSLHMGHALDHTLMDALTRRKRMQGYEVLWLPGMDH
Mvan_3951|M.vanbaalenii_PYR-1 KPAYSIVLPPPNVTGSLHMGHALDHTLMDALTRRKRMQGYEVLWLPGMDH
MSMEG_4630|M.smegmatis_MC2_155 KPPYSIVLPPPNVTGSLHMGHALDHTLMDVLTRRKRMQGYEVLWLPGMDH
TH_0609|M.thermoresistible__bu KPSYSIVLPPPNVTGSLHMGHALDHTLMDALTRRKRMQGYEVLWLPGMDH
MAB_1603|M.abscessus_ATCC_1997 KPGYSIVLPPPNVTGSLHMGHALDHTLMDALTRRKRMQGYEVLWLPGMDH
Mb2475c|M.bovis_AF2122/97 KPAYSIVLPPPNVTGSLHMGHALEHTMMDALTRRKRMQGYEVLWQPGTDH
Rv2448c|M.tuberculosis_H37Rv KPAYSIVLPPPNVTGSLHMGHALEHTMMDALTRRKRMQGYEVLWQPGTDH
MMAR_3772|M.marinum_M KPPYSIVLPPPNVTGSLHMGHALEHTMMDALTRRKRMQGYEVLWQPGMDH
MUL_3719|M.ulcerans_Agy99 KPPYSIVLPPPNVTGSLHMGHALEHTMMDALTRRKRMQGYEVLWQPGMDH
MLBr_01472|M.leprae_Br4923 KPGYSIVLPPPNVTGSLHMGHALEHTMMDALTRRKRMQGYEVLWQPGMDH
MAV_1724|M.avium_104 KPGYSIVLPPPNVTGSLHMGHALEHTMMDALTRRKRMQGYEVLWQPGMDH
** ********************:**:**.************** ** **
Mflv_2630|M.gilvum_PYR-GCK AGIATQSVVEKQLAADGKTKEDFGRELFIDKVWDWKRESGGTIGAQMRRI
Mvan_3951|M.vanbaalenii_PYR-1 AGIATQTVVEKQLSADGKTKEDFGRELFIDKVWDWKRESGGTIGGQMRRL
MSMEG_4630|M.smegmatis_MC2_155 AGIATQTVVEKQLAADGKTKEDLGREEFVAKVWDWKRESGGTIGGQMRRL
TH_0609|M.thermoresistible__bu AGIATQTVVEKQLAAQGIRKEDLGREKFIEKVWEWKRQSGGTIGAQMRRL
MAB_1603|M.abscessus_ATCC_1997 AGIATQSVVEKQLAADGKTKEDFGRELFIEKVWDWKRESGGTIGGQMRRL
Mb2475c|M.bovis_AF2122/97 AGIATQSVVEQQLAVDGKTKEDLGRELFVDKVWDWKRESGGAIGGQMRRL
Rv2448c|M.tuberculosis_H37Rv AGIATQSVVEQQLAVDGKTKEDLGRELFVDKVWDWKRESGGAIGGQMRRL
MMAR_3772|M.marinum_M AGIATQTVVEKQLAADGKTKEDFGRELFVDKVWDWKRESGGAIGGQMRRL
MUL_3719|M.ulcerans_Agy99 AGIATQTVVEKQLAADGKTKEDFGRELFVDKVWDWKRESGGAIGGQMRRL
MLBr_01472|M.leprae_Br4923 AGIATQSVVEKQLAINGKIKEDFGRELFVDKVWDWKRESGGAIAGQMRRL
MAV_1724|M.avium_104 AGIATQSVVEKQLAVDGKTKEDFGRELFIEKVWDWKRESGGAIGGQMRRL
******:***:**: :* ***:*** *: ***:***:***:*..****:
Mflv_2630|M.gilvum_PYR-GCK GDGVDWSRDRFTMDDTLSRAVRTIFKRLFDAGLIYQAERLVNWSPVLETA
Mvan_3951|M.vanbaalenii_PYR-1 GDGVDWSRDRFTMDEGLSRAVRTIFKRLFDAGLIYQAERLVNWSPVLETA
MSMEG_4630|M.smegmatis_MC2_155 GDGVDWSRDRFTMDEGLSRAVRTIFKRLFDAGLIYQAERLVNWSPVLETA
TH_0609|M.thermoresistible__bu GDGVDWSRDRFTMDEGLSRAVRTIFKQLYDAGLIYQAERLVNWSPVLQTA
MAB_1603|M.abscessus_ATCC_1997 GDGVDWSRDRFTMDEGLSRAVRTIFKKLFDAGLIYRAERLVNWSPELKTA
Mb2475c|M.bovis_AF2122/97 GDGVDWSRDRFTMDEGLSRAVRTIFKRLYDAGLIYRAERLVNWSPVLQTA
Rv2448c|M.tuberculosis_H37Rv GDGVDWSRDRFTMDEGLSRAVRTIFKRLYDAGLIYRAERLVNWSPVLQTA
MMAR_3772|M.marinum_M GDGVDWSRDRFTMDEGLSRAVRTIFKRLYDAGLIYQAERLVNWSPVLQTA
MUL_3719|M.ulcerans_Agy99 GDGVDWSRDRFTMDEGLSRAVRTIFKRLYDAGLIYQAERLVNWSPVLQTA
MLBr_01472|M.leprae_Br4923 GDGVDWSRDRFTMDDGLSRAVRMIFKRLYDAGLIYRAERLVNWSPVLRTA
MAV_1724|M.avium_104 GDGVDWSRDRFTMDEGLSRAVRTIFKRLYDAGLIYRAERLVNWSPVLQTA
**************: ****** ***:*:******:********* *.**
Mflv_2630|M.gilvum_PYR-GCK ISDLEVKYEDVEGELVSFRYGSMDDSEPHIVVATTRLETMLADTAIAVHP
Mvan_3951|M.vanbaalenii_PYR-1 ISDLEVKYEDVEGELVSFRYGSMEDSEPHIVVATTRLETMLGDTAIAVHP
MSMEG_4630|M.smegmatis_MC2_155 ISDLEVKYEDVEGELVSFRYGSMNDDEPHIVVATTRVETMLGDTAIAVHP
TH_0609|M.thermoresistible__bu ISDLEVKYEDVEGELVSFRYGSMSDDEPHIVVATTRLETMLGDTAIAVHP
MAB_1603|M.abscessus_ATCC_1997 ISDLEVKYEEVEGELVSFRYGSLNDNEPHLVVATTRLETMLGDTAIAVHP
Mb2475c|M.bovis_AF2122/97 ISDLEVNYRDVEGELVSFRYGSLDDSQPHIVVATTRVETMLGDTAIAVHP
Rv2448c|M.tuberculosis_H37Rv ISDLEVNYRDVEGELVSFRYGSLDDSQPHIVVATTRVETMLGDTAIAVHP
MMAR_3772|M.marinum_M ISDLEVNYLEVEGELVSFRYGSLEDSQPHIVVATTRVETMLGDTAIAVHP
MUL_3719|M.ulcerans_Agy99 ISDLEVNYLEVEGELVSFRYGSLEDSQPHIVVATTRVETMLGDTAIAVHP
MLBr_01472|M.leprae_Br4923 LSDIEVIYDEIEGELISFRYGSLDDDEPHIVVATTRVETMLGDTGIAVHP
MAV_1724|M.avium_104 LSDIEVNYEEVEGELVSFRYGSLDDSGPHIVVATTRVETMLGDTAIAVHP
:**:** * ::****:******:.*. **:******:****.**.*****
Mflv_2630|M.gilvum_PYR-GCK EDDRYRELVGTTLPHPFLDTELIVVADSHVDPEFGTGAVKVTPAHDPNDF
Mvan_3951|M.vanbaalenii_PYR-1 DDERYRHLVGTTLPHPFIDRQIVIVADHHVDPEFGTGAVKVTPAHDPNDF
MSMEG_4630|M.smegmatis_MC2_155 DDERYRHLVGKTLPHPFVDRDMVIVADTHVDPEFGTGAVKVTPAHDPNDF
TH_0609|M.thermoresistible__bu EDERYKHLVGKTLPHPFLDRELVIVADEHVDPEFGSGAVKVTPAHDPNDF
MAB_1603|M.abscessus_ATCC_1997 DDERYRHLIGTELAHPFQDRSIPIVADNHVDPEFGTGAVKVTPAHDPNDF
Mb2475c|M.bovis_AF2122/97 DDERYRHLVGTSLAHPFVDRELAIVADEHVDPEFGTGAVKVTPAHDPNDF
Rv2448c|M.tuberculosis_H37Rv DDERYRHLVGTSLAHPFVDRELAIVADEHVDPEFGTGAVKVTPAHDPNDF
MMAR_3772|M.marinum_M DDERYRHLVGTTLAHPFVDRQLIIVADGHVDPEFGTGAVKVTPAHDPNDF
MUL_3719|M.ulcerans_Agy99 DDERYRHLVGTTLAHPFVDRQLIIVADGHVDPEFGTGAVKVTPAHDPNDF
MLBr_01472|M.leprae_Br4923 DDKRYQHLVGTTLPHPFIDRELVIVADEHVDPEFGTGAVKVTPAHDPNDF
MAV_1724|M.avium_104 DDERYRHLVGSSLPHPFVDRQLLIVADEHVDPEFGTGAVKVTPAHDPNDF
:*.**:.*:*. *.*** * .: :*** *******:**************
Mflv_2630|M.gilvum_PYR-GCK EIGLRHQLPMPTMLETRGRVAGTGTQFDGMDRFEARVAVREALAEQGRIV
Mvan_3951|M.vanbaalenii_PYR-1 EIGLRHQLPMPSIMDTKGRIADTRTQFDGLDRFEARFAVRQALADEGRIV
MSMEG_4630|M.smegmatis_MC2_155 EIGMRHGLPMPTIMDTKGRIADTGTQFDGMDRFEARVKVREALAAQGRIV
TH_0609|M.thermoresistible__bu EIGMRHGLPMITIMDTRGRIADTGTQFDGMDRFEARVKVREALAEQGRIV
MAB_1603|M.abscessus_ATCC_1997 EIGLRHNLPMPTIMDVHGRICDSGTQFDGMDRFEARVKVREALAEQGRIV
Mb2475c|M.bovis_AF2122/97 EIGVRHQLPMPSILDTKGRIVDTGTRFDGMDRFEARVAVRQALAAQGRVV
Rv2448c|M.tuberculosis_H37Rv EIGVRHQLPMPSILDTKGRIVDTGTRFDGMDRFEARVAVRQALAAQGRVV
MMAR_3772|M.marinum_M EIGMRHQLPMPSILDTKGRIVDTGTQFDGMDRFEARVAVREALAAQGRVV
MUL_3719|M.ulcerans_Agy99 EIGMRHQLPMPSILDTKGRIVDTGTQFDGMDRFEARVAVREALAAQGRVV
MLBr_01472|M.leprae_Br4923 EIGLRHNLPMPNVMDVKAVIVDTGTEFDGMDRFEARIAVREALAVQGRIV
MAV_1724|M.avium_104 EIGLRHQLPMISIMDTKGRIADTGTQFDGMDRFAARVAVREALAAQGRIV
***:** *** .:::.:. : .: *.***:*** **. **:*** :**:*
Mflv_2630|M.gilvum_PYR-GCK AEKRPYLHSVGHSERSGEPIEPRLSLQWWVRVETLAKAAGDAVRNGDTVI
Mvan_3951|M.vanbaalenii_PYR-1 EEKRPYLHSVGHSERSGEPIEPRLSLQWWVKVESLAKAAGDAVRNGRTVI
MSMEG_4630|M.smegmatis_MC2_155 AEKRPYLHSVGHSERSGEPIEPRLSLQWWVKVESLAKAAGDAVRNGDTVI
TH_0609|M.thermoresistible__bu AEKRPYVHSVGHSERSGEPIEPRLSLQWWVKVADLAKAAGDAVRNGETVI
MAB_1603|M.abscessus_ATCC_1997 AEKRPYLHSVGHSERSGEPIEPRLSMQWWVKVESLARKAGDAVRSGDTVI
Mb2475c|M.bovis_AF2122/97 EEKRPYLHSVGHSERSGEPIEPRLSLQWWVRVESLAKAAGDAVRNGDTVI
Rv2448c|M.tuberculosis_H37Rv EEKRPYLHSVGHSERSGEPIEPRLSLQWWVRVESLAKAAGDAVRNGDTVI
MMAR_3772|M.marinum_M EEKRPYLHSVGHSDRSGEPIEPRLSLQWWVRVESLAKAAGDAVRNGDTVI
MUL_3719|M.ulcerans_Agy99 EEKRPYLHSVGHSDRSGEPFEPRLSLQWWVRVESLAKAAGDAVRNGDTVI
MLBr_01472|M.leprae_Br4923 EEKRPYRHSVGHSERSGEVIEPRLSLQWWVRVESLAKAAGDAVRNGDTVI
MAV_1724|M.avium_104 EEKRPYLHSVGHSERSGEPIEPRLSLQWWVRVESLAKAAGDAVRNGDTVI
***** ******:**** :*****:****:* **: ******.* ***
Mflv_2630|M.gilvum_PYR-GCK HPASLEPRWFAWVDNMHDWCISRQLWWGHRIPIWHGPNGETVCVGPDETP
Mvan_3951|M.vanbaalenii_PYR-1 HPASLEPRWFGWVDNMHDWCISRQLWWGHRIPIWHGPNGETVCVGPDETP
MSMEG_4630|M.smegmatis_MC2_155 HPPSLEPRWFAWVDNMHDWCISRQLWWGHRIPIWHGPDGETVCVGPDETP
TH_0609|M.thermoresistible__bu HPKSLEPRWFAWVDDMHDWCISRQLWWGHRIPIWHGPNGEKVCVGPDETP
MAB_1603|M.abscessus_ATCC_1997 HPASLEPRWFAWVDDMHDWCISRQLWWGHRIPIWHGPGGEIVCLGPDEAA
Mb2475c|M.bovis_AF2122/97 HPASMEPRWFSWVDDMHDWCISRQLWWGHRIPIWYGPDGEQVCVGPDETP
Rv2448c|M.tuberculosis_H37Rv HPASMEPRWFSWVDDMHDWCISRQLWWGHRIPIWYGPDGEQVCVGPDETP
MMAR_3772|M.marinum_M HPASLEPRWFAWVDDMHDWCISRQLWWGHRIPIWYGPNGEKVCVGPDETP
MUL_3719|M.ulcerans_Agy99 HPASLEPRWFAWVDDMHDWCISRQLWWGHRIPIWYGPDGEKVCVGPDETP
MLBr_01472|M.leprae_Br4923 HPASMESRWFAWVDDMRDWCISRQLWWGHRIPIWYGPNGEQVCVGPDETP
MAV_1724|M.avium_104 HPTSMEPRWFAWVDDMHDWCISRQLWWGHRIPIWYGPNGEQRCVGPDETP
** *:*.***.***:*:*****************:**.** *:****:.
Mflv_2630|M.gilvum_PYR-GCK PEGWEQDPDVLDTWFSSALWPFSTMGWPDHTPELAKFYPTTVLVTGYDIL
Mvan_3951|M.vanbaalenii_PYR-1 PEGWEQDPDVLDTWFSSALWPFSTMGWPDHTAELAKFYPTTVLVTGYDIL
MSMEG_4630|M.smegmatis_MC2_155 PEGWEQDPDVLDTWFSSALWPFSTMGWPDHTPELAKFYPTSVLVTGYDIL
TH_0609|M.thermoresistible__bu PEGWEQDPDVLDTWFSSALWPFSTMGWPDHTPELAKFYPTDVLVTGYDIL
MAB_1603|M.abscessus_ATCC_1997 PEGYEQDPDVLDTWFSSALWPFSTMGWPEATPELEKFYPTSVLVTGYDIL
Mb2475c|M.bovis_AF2122/97 PQGWEQDPDVLDTWFSSALWPFSTLGWPDKTAELEKFYPTSVLVTGYDIL
Rv2448c|M.tuberculosis_H37Rv PQGWEQDPDVLDTWFSSALWPFSTLGWPDKTAELEKFYPTSVLVTGYDIL
MMAR_3772|M.marinum_M PQGWEQDSDVLDTWFSSALWPFSTLGWPELTPELEKFYPTSVLVTGYDIL
MUL_3719|M.ulcerans_Agy99 PQGWEQDSDVLDTWFSSALWPFSTLGWPELTPELEKFYPTSVLVTGYDIL
MLBr_01472|M.leprae_Br4923 PEGWQQDPDVLDTWFSSALWPFSTLGWPQMTPELEKFYPTSILVTGYDIL
MAV_1724|M.avium_104 PEGWEQDPDVLDTWFSSALWPFSTLGWPEKTPELEKFYPTSVLVTGYDIL
*:*::**.****************:***: *.** ***** :********
Mflv_2630|M.gilvum_PYR-GCK FFWVARMMMFGTFVGGDPAIIGDGAAKKSSPQVPFKNVFLHGLIRDEFGR
Mvan_3951|M.vanbaalenii_PYR-1 FFWVARMMMFGTFVGDDPAITGGGAR---GPQVPFENVFLHGLIRDEHGR
MSMEG_4630|M.smegmatis_MC2_155 FFWVARMMMFGTFVGDDPAVTLEGAR---GRQVPFENVFLHGLIRDEFGR
TH_0609|M.thermoresistible__bu FFWVARMMMFGTFVGDHPAITLDGAR---GPQVPFRNVFLHGLIRDKHGR
MAB_1603|M.abscessus_ATCC_1997 FFWVARMMMFGTFVADGPALH--------GGKVPFDNVFLHGLIRDQFGQ
Mb2475c|M.bovis_AF2122/97 FFWVARMMMFGTFVGDDAAITLDGRR---GPQVPFTDVFLHGLIRDESGR
Rv2448c|M.tuberculosis_H37Rv FFWVARMMMFGTFVGDDAAITLDGRR---GPQVPFTDVFLHGLIRDESGR
MMAR_3772|M.marinum_M FFWVARMMMFGTFVGGDEVITLDGRR---GAQVPFTDVFLHGLIRDEFGR
MUL_3719|M.ulcerans_Agy99 FFWVARMMMFGTFVGGDEVITLDGRR---GAQVPFTDVFLHGLIRDEFGR
MLBr_01472|M.leprae_Br4923 FFWVARMMMLGTFVGGDDAITLGGCR---GPQVPFTDVFLHGLIRDEFGR
MAV_1724|M.avium_104 FFWVARMMMFGTFVGDDDAITLDGRR---GPQVPFTDVFLHGLIRDESGR
*********:****.. .: . :*** :*********: *:
Mflv_2630|M.gilvum_PYR-GCK KMSKSRGNGIDPLDWVETFGADALRFTLARGASPGGDLAIGEDHARASRN
Mvan_3951|M.vanbaalenii_PYR-1 KMSKSRGNGIDPLDWVEMFGADALRFTLARGASPGGDLAIGEDHARASRN
MSMEG_4630|M.smegmatis_MC2_155 KMSKSRGNGIDPLDWVEKFGADALRFTLARGASPGGDLSIGEDHARASRN
TH_0609|M.thermoresistible__bu KMSKSAGNGIDPLDWVQTYGADALRFTLARGASPGSDLSIGDDHARASRN
MAB_1603|M.abscessus_ATCC_1997 KMSKSKGNGIDPLDWVEMFGADALRFTLARGASPGGDLSIGEDAARASRN
Mb2475c|M.bovis_AF2122/97 KMSKSKGNVIDPLDWVEMFGADALRFTLARGASPGGDLAVSEDAVRASRN
Rv2448c|M.tuberculosis_H37Rv KMSKSKGNVIDPLDWVEMFGADALRFTLARGASPGGDLAVSEDAVRASRN
MMAR_3772|M.marinum_M KMSKSKGNVIDPLDWIDTFGADALRFTLARGASPGSDLAVGEDAVRASRN
MUL_3719|M.ulcerans_Agy99 KMSKSKGNVIDPLDWIDTFGADALRFTLARGASPGSDLAVGEDAVRASRN
MLBr_01472|M.leprae_Br4923 KMSKSRGNVIDPLAWMDMFGADALRFTLARGSSPGGDLAIGEDHVRASRN
MAV_1724|M.avium_104 KMSKSKGNVIDPLDWVDMFGADALRFTLARGASPGGDLAIGEDHVRASRN
***** ** **** *:: :************:***.**::.:* .*****
Mflv_2630|M.gilvum_PYR-GCK FATKLFNATRFALLNGASPAPLPPTAELTDADRWILGRLEEVRADVDAAL
Mvan_3951|M.vanbaalenii_PYR-1 FATKLFNATRFALLNGASPAPLPPSDQLTDADRWILGRLEEVRIEVDAAL
MSMEG_4630|M.smegmatis_MC2_155 FATKLFNATRFALMNGAAPAPLPAASELTDADRWILGRLEEVRAEVDSAF
TH_0609|M.thermoresistible__bu FVTKLFNATRFALLNGAAPAPLPDAAELTDADRWILGRLEEVRTEVDSAF
MAB_1603|M.abscessus_ATCC_1997 FTTKLFNATRFALMNGAALGELPDVAELTDADRWILGRLEQVRAEVDGAL
Mb2475c|M.bovis_AF2122/97 FGTKLFNATRYALLNGAAPAPLPSPNELTDADRWILGRLEEVRAEVDSAF
Rv2448c|M.tuberculosis_H37Rv FGTKLFNATRYALLNGAAPAPLPSPNELTDADRWILGRLEEVRAEVDSAF
MMAR_3772|M.marinum_M FGTKLFNATRYALLNGAALAPLPPLGELTDADRWILGRLEEVRAEVDSAF
MUL_3719|M.ulcerans_Agy99 FGTKLFNATRYALLNGAALAPLPPLGELTDADRWILGRLEEVRAEVDSAF
MLBr_01472|M.leprae_Br4923 FGTKLFNATRYALLNGAALVPLPALTALTDADRWILGRLEQVRAEVDSAF
MAV_1724|M.avium_104 FCTKLFNATRYALLNGAQLAQLPPLDELTDADRWILGRLEEVRAEVDSAF
* ********:**:*** ** *************:** :**.*:
Mflv_2630|M.gilvum_PYR-GCK DNYEFNRACEALYHFAWDEFCDWYLELAKVQLGQEDPQTARTTAVLATVL
Mvan_3951|M.vanbaalenii_PYR-1 DNYEFSRACESLYHFAWDEFCDWYLELAKVQLGQEEAHTARTTAVLAFAL
MSMEG_4630|M.smegmatis_MC2_155 DNYEFNRACEALYHFAWDEFCDWYVELAKVQLGQEEVHTARTTAVLAAVL
TH_0609|M.thermoresistible__bu DSYEFSRACETLYHFAWDEFCDWYVELAKVQLSEG---ISHTTAVLAAVL
MAB_1603|M.abscessus_ATCC_1997 DRYEFSVACEGLYHFAWDEFCDWYLELAKVQMVDR---AESTRVVLATVL
Mb2475c|M.bovis_AF2122/97 DGYEFSRACESLYHFAWDEFCDWYLELAKTQLAQG---LTHTTAVLAAGL
Rv2448c|M.tuberculosis_H37Rv DGYEFSRACESLYHFAWDEFCDWYLELAKTQLAQG---LTHTTAVLAAGL
MMAR_3772|M.marinum_M DSYEFSRACEALYHFAWDEFCDWYVELAKTQLAEG---LSATTAVLAAGL
MUL_3719|M.ulcerans_Agy99 DSYEFSRACEALYHFAWDEFCDWYVELAKTQLAEG---LSATTAVLAAGL
MLBr_01472|M.leprae_Br4923 DGYEFSRACEALYHFAWDEFCDWYLELAKAQLADG---LTHTTAVLAAAL
MAV_1724|M.avium_104 DNYEFSRACESLYHFAWDEFCDWYVELAKTQLAEG---ITHTTAVLATTL
* ***. *** *************:****.*: : * .*** *
Mflv_2630|M.gilvum_PYR-GCK DVLLKLLHPVMPFVTEVLWKTLTGGESLVIADWPTATGFASDHAAAQRIT
Mvan_3951|M.vanbaalenii_PYR-1 DVLLKLLHPVMPFVTEVLWKALTGGESLVIADWPTATGYAVDQGATQRVA
MSMEG_4630|M.smegmatis_MC2_155 DTLLKLLHPVMPFVTETLWKTLTGGESVVVATWPEPSGFAPDTVATQRIT
TH_0609|M.thermoresistible__bu DTLLKLLHPVMPFVTEKLWKTLTDGESLVIAKWPQPSGFSLDSIAAQRVA
MAB_1603|M.abscessus_ATCC_1997 DTLLRLLHPVIPFVTEVLWKSVTGGESVVIAAWPTGSGIELDGTAAQRIA
Mb2475c|M.bovis_AF2122/97 DTLLRLLHPVIPFLTEALWLALTGRESLVSADWPEPSGISVDLVAAQRIN
Rv2448c|M.tuberculosis_H37Rv DTLLRLLHPVIPFLTEALWLALTGRESLVSADWPEPSGISVDLVAAQRIN
MMAR_3772|M.marinum_M DTLLRLLHPVIPFITEALWQALTGQESLVIADWPQPSGITLDPVAAQRIS
MUL_3719|M.ulcerans_Agy99 DTLLRLLHPVIPFITEALWQALTGQESLVIADWPQPSGITLDPVAAQRIS
MLBr_01472|M.leprae_Br4923 DTLLRLLHPVMPFITETLWQALTQLESLVIATWPEPSGISLDLVAAQRIS
MAV_1724|M.avium_104 DTLLRLLHPVIPFITEALWQALTGNESLVIADWPRSSGVGLDQVATQRIT
*.**:*****:**:** ** ::* **:* * ** :* * *:**:
Mflv_2630|M.gilvum_PYR-GCK DMQKLITEVRRFRSDQGLADRQKVPARLSEVSAAGLDAHVPAVTALAWLT
Mvan_3951|M.vanbaalenii_PYR-1 DMQKLITEVRRFRSDQGLADRQKVPARLSDVAAAGLEEHVPAVTALAWLT
MSMEG_4630|M.smegmatis_MC2_155 DMQKLITEVRRFRSDQGLADRQKVPARLTDVEAAGLTAQLPAVAALAWLT
TH_0609|M.thermoresistible__bu DMQRLITEVRRFRSDQGLADRQKVPARLSDIPGADLQAQLPAIRALAWLT
MAB_1603|M.abscessus_ATCC_1997 DMQKLVTEVRRFRSDQGLGDRQKVAARLSGVGEAGLDDQLAAVTSLAWLT
Mb2475c|M.bovis_AF2122/97 DMQKLVTEVRRFRSDQGLADRQKVPARMHGVRDSDLSNQVAAVTSLAWLT
Rv2448c|M.tuberculosis_H37Rv DMQKLVTEVRRFRSDQGLADRQKVPARMHGVRDSDLSNQVAAVTSLAWLT
MMAR_3772|M.marinum_M DMQNLVTEIRRFRSDQGLADRQKVPARLIGVEESDLGAQVAAVTSLAWLT
MUL_3719|M.ulcerans_Agy99 DMQNLVTEIRRFRSDQGLADRQKVPARLIGVEESDLGAQVAAVTSLAWLT
MLBr_01472|M.leprae_Br4923 DMQKLVTEIRRFRSDQGLVDRQKVPARLSGVEDSDLATQVGFVTSLALLT
MAV_1724|M.avium_104 DMQKLVTEVRRFRSDQGLADRQKVPARLAGVTESDLDTQVSAVTSLAWLT
***.*:**:********* *****.**: : :.* :: : :** **
Mflv_2630|M.gilvum_PYR-GCK PPAEGFTPSASVEVRLSAS---TVVVEVDTSGTVDVDAERRRLEKDLAAA
Mvan_3951|M.vanbaalenii_PYR-1 PPADEFTPSAAVEVRLSTA---TVVVEVDTSGTVDVAAERRRLEKDLAAA
MSMEG_4630|M.smegmatis_MC2_155 EAGDGFSPSASVEVRLSQG---TVVVEVDTSGTVDVEAERRRLEKDLGGR
TH_0609|M.thermoresistible__bu DAGADFNPSAALEVRLSKG---TVVVELDTRGSVDVEAERRRTEKDLAAA
MAB_1603|M.abscessus_ATCC_1997 PAEEGFSPTASVEVRLSGA---TVTVEVDTSGTVDVEAERRRLEKDLAAA
Mb2475c|M.bovis_AF2122/97 EPGPDFEPSVSLEVRLGPEMNRTVVVELDTSGTIDVAAERRRLEKELAGA
Rv2448c|M.tuberculosis_H37Rv EPGPDFEPSVSLEVRLGPEMNRTVVVELDTSGTIDVAAERRRLEKELAGA
MMAR_3772|M.marinum_M APGEGFAPSVSLEVRLGPSMNHTVVVELDTSGTIDVAAERRRLEKDLAAA
MUL_3719|M.ulcerans_Agy99 APGEGFAPSVSLEVRLGPSMNHTVVVELDTSGTIDVAAERRRLEKDLAAA
MLBr_01472|M.leprae_Br4923 AASNDFRPSALLEVRLGPNKDRAVVVELDTSGTIDVAAERRRMEKDLAAA
MAV_1724|M.avium_104 DAGPDFRPSASVEVRLRGG---TVVVELDTSGSIDVAAERRRLEKDLAAA
. * *:. :**** :*.**:** *::** ***** **:*..
Mflv_2630|M.gilvum_PYR-GCK QKELAGTSAKLGNDAFLAKAPADVVDKIRARRQVAGEEVDRITARLSALP
Mvan_3951|M.vanbaalenii_PYR-1 QKELSGTSGKLGNEAFLAKAPADVVDKIRARQQVANEEVERIQARLAALS
MSMEG_4630|M.smegmatis_MC2_155 PEGTGHHHGQTRQRGLPVQG-----ARPGGRQDPGQASPGQ---------
TH_0609|M.thermoresistible__bu RRELESTSKKLENEAFLAKAPAEVVEKIRNRQRLAAEEVDRLTARLAALK
MAB_1603|M.abscessus_ATCC_1997 RKELDGTTSKLGNDAFLAKAPEAVVEKIRVRQQVATEEVERITARLAALA
Mb2475c|M.bovis_AF2122/97 QKELASTAAKLANADFLAKAPDAVIAKIRDRQRVAQQETERITTRLAALQ
Rv2448c|M.tuberculosis_H37Rv QKELASTAAKLANADFLAKAPDAVIAKIRDRQRVAQQETERITTRLAALQ
MMAR_3772|M.marinum_M EKELVSTAAKLANADFLAKAPEAVVGKIRDRQRLAQEESDRITTRLAALQ
MUL_3719|M.ulcerans_Agy99 EKELVSAAAKLANADFLAKAPEAVVGKIRDRQRLAQEESDRITTRLAALQ
MLBr_01472|M.leprae_Br4923 QKELASTAAKLANADFLAKAPEAVVVKIRDRQRMAKEETDRIIARLAGLQ
MAV_1724|M.avium_104 HKELASTTAKLANEDFLAKAPPHVVDKIRDRQRLAQEESERINARLAVLQ
. : : : .:. : *: . . :
Mflv_2630|M.gilvum_PYR-GCK AGE
Mvan_3951|M.vanbaalenii_PYR-1 ASE
MSMEG_4630|M.smegmatis_MC2_155 ---
TH_0609|M.thermoresistible__bu DS-
MAB_1603|M.abscessus_ATCC_1997 ---
Mb2475c|M.bovis_AF2122/97 ---
Rv2448c|M.tuberculosis_H37Rv ---
MMAR_3772|M.marinum_M ---
MUL_3719|M.ulcerans_Agy99 ---
MLBr_01472|M.leprae_Br4923 ---
MAV_1724|M.avium_104 ---