For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RPFYVTTAITYPNGDPHVGHAYEYIATDAIARFKRLDGFDVRFLTGTDEHGLKMVETAAAEGIGTAELAR RNSDVFQRLQERLNISFDRFIRTTDPDHHEASKAIWRRMQAAGDIYLDSYSGWYSVRDERFFVESETKVL DDGTRVAAETGAPLTWTEEQTYFFRLSAYTDKLLAHYEANPDFIAPEVRRNEVVSFVSGGLRDLSISRTS FDWGVKVPDHPDHVMYVWVDALTNYLTGVGYPDTESEMFRRYWPADLHMIGKDIIRFHTVYWPAFLMSAG IELPRRVFAHGFLLNRGEKMSKSVGNIVDPITLVDTFGVDQLRYFLLREVPFGQDGSYTKDAIITRINTD LANELGNLAQRSLSMIAKNLHSVVPEPGDFTGDDRELLQTADGLLPRVRASFDSQAMHLGLEAIWLMLGE ANKYFSAQQPWVLRKSESEADQTRFRTVLYVTCEVVRIAALLVQPVMPDSAGKLLDLLGQGQDRRAFDAL GARLAPGTVLPPPTGVFPRYQAESEKVD*
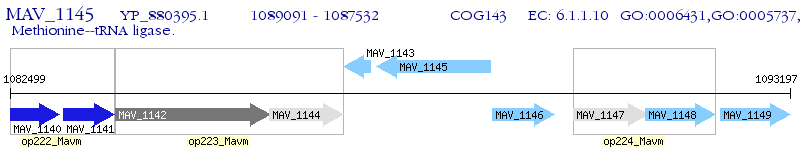
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_1145 | metG | - | 100% (519) | methionyl-tRNA synthetase |
| M. avium 104 | MAV_1724 | valS | 1e-11 | 27.70% (213) | valyl-tRNA synthetase |
| M. avium 104 | MAV_0055 | leuS | 5e-08 | 31.45% (124) | leucyl-tRNA synthetase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1034c | metG | 0.0 | 82.81% (512) | methionyl-tRNA synthetase |
| M. gilvum PYR-GCK | Mflv_1930 | - | 0.0 | 78.47% (511) | methionyl-tRNA synthetase |
| M. tuberculosis H37Rv | Rv1007c | metG | 0.0 | 82.62% (512) | methionyl-tRNA synthetase |
| M. leprae Br4923 | MLBr_00238 | metS | 0.0 | 80.81% (521) | methionyl-tRNA synthetase |
| M. abscessus ATCC 19977 | MAB_1128c | - | 0.0 | 75.88% (514) | methionyl-tRNA synthetase |
| M. marinum M | MMAR_4481 | metS | 0.0 | 83.40% (512) | methionyl-tRNA synthetase MetS |
| M. smegmatis MC2 155 | MSMEG_5441 | metG | 0.0 | 78.24% (510) | methionyl-tRNA synthetase |
| M. thermoresistible (build 8) | TH_1139 | metS | 0.0 | 81.45% (512) | PROBABLE METHIONYL-TRNA SYNTHETASE METS (MetRS) |
| M. ulcerans Agy99 | MUL_4654 | metS | 0.0 | 82.81% (512) | methionyl-tRNA synthetase |
| M. vanbaalenii PYR-1 | Mvan_4803 | - | 0.0 | 79.30% (512) | methionyl-tRNA synthetase |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_1128c|M.abscessus_ATCC_199 MPESLDPFYITTAIAYPNGEPHIGHAYEYVATDAVARFKRLDGFDVRYLT
MSMEG_5441|M.smegmatis_MC2_155 ---MSEPFYITTAIAYPNGVPHIGHAYEYIATDAIARFKRLDGYDVRYLT
Mflv_1930|M.gilvum_PYR-GCK ---MSQPYYVTTAIAYPNGDPHVGHAYEYIATDAIARFKRLDGFDVRFLT
Mvan_4803|M.vanbaalenii_PYR-1 ---MTEPYYITTAIAYPNGDPHVGHAYEYIATDAVARFKRLDGFDVRFLT
TH_1139|M.thermoresistible__bu ---MSEPYYLTTAIAYPNGDPHVGHAYEYIASDAIARFKTLDGYDVRFLT
Mb1034c|M.bovis_AF2122/97 ----MKPYYVTTAIAYPNAAPHVGHAYEYIATDAIARFKRLDGYDVRFLT
Rv1007c|M.tuberculosis_H37Rv ----MKPYYVTTAIAYPNAAPHVGHAYEYIATDAIARFKRLDRYDVRFLT
MLBr_00238|M.leprae_Br4923 ----MRPYYITTAIAYPNAAPHIGHAYEYIATDAIARFKRLDGLDVRFLT
MMAR_4481|M.marinum_M ----MKPYYVTTAIAYPNAAPHVGHAYEYIATDAIARFKRLDGFDVRFLT
MUL_4654|M.ulcerans_Agy99 ----MKPYYVTTAIAYPNAAPHVGHAYEYIATDAIARFKRLDGFDVRFLT
MAV_1145|M.avium_104 ----MRPFYVTTAITYPNGDPHVGHAYEYIATDAIARFKRLDGFDVRFLT
*:*:****:***. **:******:*:**:**** ** ***:**
MAB_1128c|M.abscessus_ATCC_199 GTDEHGLKVAQTAAAAGIPTAQLARHNSDVFEALQRTLNISFDRFIRTTD
MSMEG_5441|M.smegmatis_MC2_155 GTDVHGQKMAETAAKEGIPAAELARRNSDVFQRLQEKLNISFDRFIRTSD
Mflv_1930|M.gilvum_PYR-GCK GTDVHGLKMAETAARLGVPTAELARSNSDVFQRLQEKLNVSFDRFIRTSD
Mvan_4803|M.vanbaalenii_PYR-1 GTDVHGLKMAETAAKLGVPTAELARSNSDVFQRLQEKLHISFDRFIRTSD
TH_1139|M.thermoresistible__bu GTDEHGQKMAETAAAEGIPTAELARRNSDKFQRLQEKLNISFDRFIRTTD
Mb1034c|M.bovis_AF2122/97 GTDEHGLKVAQAAAAAGVPTAALARRNSDVFQRMQEALNISFDRFIRTTD
Rv1007c|M.tuberculosis_H37Rv GTDEHGLKVAQAAAAAGVPTAALARRNSDVFQRMQEALNISFDRFIRTTD
MLBr_00238|M.leprae_Br4923 GTDEHGLKVAQAAEAAGVPTAQLARRNSGVFQRMQEALHISFDRFIRTTD
MMAR_4481|M.marinum_M GTDEHGLKVAQAAAAAEIPTAQLARRNSDVFQRMQEALNISFDRFIRTTD
MUL_4654|M.ulcerans_Agy99 GTDEHGLKVAQAAAAADIPTAQLARRNSDVFQRMQEALNISFDRFIRTTD
MAV_1145|M.avium_104 GTDEHGLKMVETAAAEGIGTAELARRNSDVFQRLQERLNISFDRFIRTTD
*** ** *:.::* : :* *** **. *: :*. *::********:*
MAB_1128c|M.abscessus_ATCC_199 PDHYEASKAIWRKMADNGDIYLDTYSGWYSVRDEAYYTDAETTVAPDGTR
MSMEG_5441|M.smegmatis_MC2_155 ADHYEASKAIWKRMADAGDIYLDAYKGWYSIRDERFFTENETTEQPDGTR
Mflv_1930|M.gilvum_PYR-GCK ADHYEASKAIWTRMVDAGDVYLDKYSGWYSVRDERFFTEAETSIGADGVR
Mvan_4803|M.vanbaalenii_PYR-1 PDHYEASKAIWRRMVDAGDVYLGKYSGWYSVRDERFFTEAETSVDAAGVR
TH_1139|M.thermoresistible__bu PDHIEASRAIWRRMNDAGDIYLDTYKGWYSVRDERFFTDAETTVRDDGSR
Mb1034c|M.bovis_AF2122/97 ADHHEASKELWRRMSAAGDIYLDNYSGWYSVRDERFFVESETQLVD-GTR
Rv1007c|M.tuberculosis_H37Rv ADHHEASKELWRRMSAAGDIYLDNYSGWYSVRDERFFVESETQLVD-GTR
MLBr_00238|M.leprae_Br4923 ADHYKAAKEIWRRMDAAGDIYLGTYSGWYSVRDERFFVDSETKLLDNGIR
MMAR_4481|M.marinum_M ADHYEASQEIWRRMDAAGDIYLDSYSGWYSVRDERFFVESETQVVD-GTR
MUL_4654|M.ulcerans_Agy99 SDHYEASQEIWRRMDAAGDIYLDSYSGWYSVRDERFFVESETQVVD-GTR
MAV_1145|M.avium_104 PDHHEASKAIWRRMQAAGDIYLDSYSGWYSVRDERFFVESETKVLDDGTR
.** :*:: :* :* **:**. *.****:*** ::.: ** * *
MAB_1128c|M.abscessus_ATCC_199 TATATGTPVDWTE-EHSYFFRLSAYQDKLLAHYEAHPEFIGPDVRRNEVV
MSMEG_5441|M.smegmatis_MC2_155 IATETGAPVTWTE-EQTYFFRLSAYTDRLLALYEEHPEFIGPDARRNEIV
Mflv_1930|M.gilvum_PYR-GCK IATETGTPVTWTE-EETYFFRLSAYTDRLLALYAERPDFIEPEVRRNEIV
Mvan_4803|M.vanbaalenii_PYR-1 IATETGTPVTWTE-EETYFFRLSAYADRLLALYADHPEFIEPEVRRNEVV
TH_1139|M.thermoresistible__bu VATETGAPVTWTE-EPTYFFRLSAYTDRLLAHYEAHPEFIGPEVRRNEVI
Mb1034c|M.bovis_AF2122/97 LTVETGTPVTWTE-EQTYFFRLSAYTDKLLAHYHANPDFIAPETRRNEVI
Rv1007c|M.tuberculosis_H37Rv LTVETGTPVTWTE-EQTYFFRLSAYTDKLLAHYHANPDFIAPETRRNEVI
MLBr_00238|M.leprae_Br4923 VAVETGTPVTWTEKEQTYFFRLSAYVDKLLAHYDANPDFIGPEVRRNEVI
MMAR_4481|M.marinum_M IATETGTPVTWTE-EQTYFFRLSAYADKLLAHYEANPDFIAPEVRRNEVV
MUL_4654|M.ulcerans_Agy99 IATETGNPVTWTE-EQTYFFRLSAYADKLLAHYEANPDFIAPEVRRNEVV
MAV_1145|M.avium_104 VAAETGAPLTWTE-EQTYFFRLSAYTDKLLAHYEANPDFIAPEVRRNEVV
:. ** *: *** * :******** *:*** * .*:** *:.****::
MAB_1128c|M.abscessus_ATCC_199 SFVSGGLRDLSISRTTFDWGVPVPDNPEHVMYVWVDALTNYLTGVGYPDT
MSMEG_5441|M.smegmatis_MC2_155 SFVSGGLKDLSISRTTFDWGVPVPDHPDHVMYVWVDALTNYLTGVGFPDT
Mflv_1930|M.gilvum_PYR-GCK SFVSGGLRDLSISRTTFDWGVPVPGHPDHVMYVWVDALTNYLTGVGFPDT
Mvan_4803|M.vanbaalenii_PYR-1 SFVSGGLRDLSISRTTFDWGVPVPDHPDHVMYVWVDALTNYLTGVGFPDT
TH_1139|M.thermoresistible__bu SFVSQGLRDLSISRTTFDWGVPVPDHPEHVMYVWVDALTNYLTGVGFPDT
Mb1034c|M.bovis_AF2122/97 SFVSGGLDDLSISRTSFDWGVQVPEHPDHVMYVWVDALTNYLTGAGFPDT
Rv1007c|M.tuberculosis_H37Rv SFVSGGLDDLSISRTSFDWGVQVPEHPDHVMYVWVDALTNYLTGAGFPDT
MLBr_00238|M.leprae_Br4923 SFVSGGLEDFSISRTSFDWGVQVPEHPDHVMYVWIDALTNYLTGAGFPDT
MMAR_4481|M.marinum_M SFVSGGLKDLSVSRTSFNWGVPVPGHPDHVMYVWVDALTNYLTGAGYPDT
MUL_4654|M.ulcerans_Agy99 SFVSGGLKDLSVSRTSFNWGVPVPGHPDHVMYVWVDALTNYLTGAGYPDT
MAV_1145|M.avium_104 SFVSGGLRDLSISRTSFDWGVKVPDHPDHVMYVWVDALTNYLTGVGYPDT
**** ** *:*:***:*:*** ** :*:******:*********.*:***
MAB_1128c|M.abscessus_ATCC_199 DSELFGRYWPANIHMIGKDIIRFHTVYWPAFLMSAGIALPQRVFAHGFLT
MSMEG_5441|M.smegmatis_MC2_155 ESESFRRYWPADLHMIGKDIIRFHTVYWPAFLMSAGLPLPKRIFAHGWLL
Mflv_1930|M.gilvum_PYR-GCK DSETFRRYWPADLHMIGKDIIRFHAVYWPAFLMSAGIELPKKVFAHGFLF
Mvan_4803|M.vanbaalenii_PYR-1 DSELFTRYWPADLHMVGKDIIRFHTVYWPAFLMSAGIELPKKVFVHGFLL
TH_1139|M.thermoresistible__bu ESELFRRYWPADLHMIGKDIIRFHTVYWPAFLMSAGLELPRRVFAHGFLY
Mb1034c|M.bovis_AF2122/97 DSELFRRYWPADLHMIGKDIIRFHAVYWPAFLMSAGIELPRRIFAHGFLH
Rv1007c|M.tuberculosis_H37Rv DSELFRRYWPADLHMIGKDIIRFHAVYWPAFLMSAGIELPRRIFAHGFLH
MLBr_00238|M.leprae_Br4923 DSELFGRYWPANLHMIGKDIIRFHAVYWPAFLMSAGIELPRRIFAHGFLH
MMAR_4481|M.marinum_M GSEAFLRYWPADLHMIGKDIIRFHAVYWPAFLMSAGIELPRRVFAHGFLH
MUL_4654|M.ulcerans_Agy99 GSEAFLRYWPADQHMIGKDIIRFHAVYWPAFLMSAGIELPRRVFAHGFLH
MAV_1145|M.avium_104 ESEMFRRYWPADLHMIGKDIIRFHTVYWPAFLMSAGIELPRRVFAHGFLL
** * *****: **:********:***********: **:::*.**:*
MAB_1128c|M.abscessus_ATCC_199 NKGEKMSKSIGNVVDPINLVNTFGLDQVRYFLLREVPFGQDGSYTEEGLI
MSMEG_5441|M.smegmatis_MC2_155 NRGEKMSKSIGNVVDPVNLVDTFGLDQVRYFLLREVPFGQDGSYNEDAII
Mflv_1930|M.gilvum_PYR-GCK NRGEKMSKSVGNVVDPNDLVETFGVDQLRYFLLREVSFGQDGSYSEDGII
Mvan_4803|M.vanbaalenii_PYR-1 NRGEKMSKSVGNVVDPVNLVDTFGLDQVRYFFLREVPFGQDGSYSEDAII
TH_1139|M.thermoresistible__bu NRGEKMSKSVGNVIDPNDLVDTFGVDQVRYFLLREVPFGQDGSYSEEAII
Mb1034c|M.bovis_AF2122/97 NRGEKMSKSVGNIVDPVALAEALGVDQVRYFLLREVPFGQDGSYSDEAIV
Rv1007c|M.tuberculosis_H37Rv NRGEKMSKSVGNIVDPVALAEALGVDQVRYFLLREVPFGQDGSYSDEAIV
MLBr_00238|M.leprae_Br4923 NHGEKMSKSVGNIVDPMALVQTFGVDQVRYFLLREIPFGQDGNYSEEAII
MMAR_4481|M.marinum_M NRGEKMSKSLGNTIDPMGLIETYGVDQVRYFLLREVPFGQDGSYSEEAII
MUL_4654|M.ulcerans_Agy99 NRGEKMSKSLGNTIDPMGLIETYGVDQVRYFLLREVPFGQDGSYSEEAII
MAV_1145|M.avium_104 NRGEKMSKSVGNIVDPITLVDTFGVDQLRYFLLREVPFGQDGSYTKDAII
*:*******:** :** * :: *:**:***:***:.*****.*..:.::
MAB_1128c|M.abscessus_ATCC_199 GRINADLANELGNLAQRSLSMVAKNLGGIVPEPGEFAPEDTELLAAADTL
MSMEG_5441|M.smegmatis_MC2_155 GRVNADLANELGNLAQRSLSMVAKNLGAAVPDPGEFTDEDTALLAAADAL
Mflv_1930|M.gilvum_PYR-GCK ARINADLANEFGNLAQRSLSMVAKNLDGAVPEPGEFTAEDTAMLAAADDL
Mvan_4803|M.vanbaalenii_PYR-1 GRINADLANEFGNLAQRSLSMVAKNLDGVVPQPGAFTAEDSEMLAAADAL
TH_1139|M.thermoresistible__bu SRINADLANDLGNLAQRSLSMVAKNLGGVVPEPGPFTDDDSALLDAADGL
Mb1034c|M.bovis_AF2122/97 TRINTDLANELGNLAQRSLSMVAKNLDGRVPNPGEFADADAALLATADGL
Rv1007c|M.tuberculosis_H37Rv TRINTDLANELGNLAQRSLSMVAKNLDGRVPNPGEFADADAALLATADGL
MLBr_00238|M.leprae_Br4923 TRMNTDLANEFGNLAQRSLSMVAKNLGGVVPEPSEFTSADTALLTTADGL
MMAR_4481|M.marinum_M NRINTDLANELGNLAQRSLSMIAKNLGGAVPEPGEFSAADRELLAIADGL
MUL_4654|M.ulcerans_Agy99 NRINTDLANELGNLAQRSLSMIAKNLGGAVPEPGEFSAADRELLAIADGL
MAV_1145|M.avium_104 TRINTDLANELGNLAQRSLSMIAKNLHSVVPEPGDFTGDDRELLQTADGL
*:*:****::**********:**** . **:*. *: * :* ** *
MAB_1128c|M.abscessus_ATCC_199 LEQARTHFEVPAMHLALEAIWQVLGASNRYFSAQEPWVLAKSFQTDGSAA
MSMEG_5441|M.smegmatis_MC2_155 LERVREHFDVPAMHLALEAIWSVLGAANRYFSAQEPWVLRKS----DAAE
Mflv_1930|M.gilvum_PYR-GCK LDRVREHYDATAMHLALEAIWSVLGAANRYFSAQEPWVLRKT--------
Mvan_4803|M.vanbaalenii_PYR-1 LERVRRHFEATAMHLALEAIWSVLGAANRYFSAQEPWVLRKT--------
TH_1139|M.thermoresistible__bu LERVRRHYDDLAMHLALEAIWSVLGAANRYFSAQQPWVLRKS----ESSQ
Mb1034c|M.bovis_AF2122/97 LERVRGHFDAQAMHLALEAIWLMLGDANKYFSVQQPWVLRKS----ESEA
Rv1007c|M.tuberculosis_H37Rv LERVRGHFDAQAMHLALEAIWLMLGDANKYFSVQQPWVLRKS----ESEA
MLBr_00238|M.leprae_Br4923 LERVRGNFDGQAMNLALEAIWLMLGEANKYFSSQQPWILRKS----ESEA
MMAR_4481|M.marinum_M LERVREHFDNQSMHLALEAIWLMLGHANKYFSDQQPWVLRKS----ESEA
MUL_4654|M.ulcerans_Agy99 LERVREHFDNQSMHLALEAIWLMLGHANKYFSDQQPWVLRKS----ESET
MAV_1145|M.avium_104 LPRVRASFDSQAMHLGLEAIWLMLGEANKYFSAQQPWVLRKS----ESEA
* :.* :: :*:*.***** :** :*:*** *:**:* *:
MAB_1128c|M.abscessus_ATCC_199 DQERFGTVLYTTLEVVRIVALLVQPVMPESAAKLLDLLGQADDQRDFAAI
MSMEG_5441|M.smegmatis_MC2_155 DQQRFRTVLYTTLEVVRIASLLLQPVMPESTAKLLDLLGQPTDERDFSAI
Mflv_1930|M.gilvum_PYR-GCK DPGRFATVLYTTLEVVRIAALLSQPVMPGTMATLLDLLGQPEDRRAFSAI
Mvan_4803|M.vanbaalenii_PYR-1 DPARFATVLYTTLEVVRIAALLSQPVMPDAAAKLLDLLGQPADRRAFTAI
TH_1139|M.thermoresistible__bu DQIRFRTVLYTTLEVVRVAALLSQPVMPEATGKLLDLLGLPAEQRTFATI
Mb1034c|M.bovis_AF2122/97 DQARFRTTLYVTCEVVRIAALLIQPVMPESAGKILDLLGQAPNQRSFAAV
Rv1007c|M.tuberculosis_H37Rv DQARFRTTLYVTCEVVRIAALLIQPVMPESAGKILDLLGQAPNQRSFAAV
MLBr_00238|M.leprae_Br4923 DQARFRTVLYTTCEVVRIAALLVQPVMPESAGKMLDLLGQEEDQRAFTAV
MMAR_4481|M.marinum_M DQARFRTTLYTTCEAVRIAALLVAPVMPESAAKLLDLLGQGPDQRSFNAI
MUL_4654|M.ulcerans_Agy99 DQARFRTTLYTTCEAVRIAALLVAPVMPESAAKLLDLLGQRPDQRSFNAI
MAV_1145|M.avium_104 DQTRFRTVLYVTCEVVRIAALLVQPVMPDSAGKLLDLLGQGQDRRAFDAL
* ** *.**.* *.**:.:** **** : ..:***** :.* * ::
MAB_1128c|M.abscessus_ATCC_199 GTRLKPGVSLPAPTGVFPKYQVPK---------------------
MSMEG_5441|M.smegmatis_MC2_155 ANRIKPGTSLPAPSGIFPRYQND----------------------
Mflv_1930|M.gilvum_PYR-GCK TVRLTPGTALPAPTGVFPRYQTTRDD-------------------
Mvan_4803|M.vanbaalenii_PYR-1 GTRLAPGTALPSPTGVFPRYQAD----------------------
TH_1139|M.thermoresistible__bu GIRITPGTALPAPVGVFPRYQAE----------------------
Mb1034c|M.bovis_AF2122/97 GVRLTPGTALPPPTGVFPRYQP-------PQPPEGK---------
Rv1007c|M.tuberculosis_H37Rv GVRLTPGTALPPPTGVFPRYQP-------PQPPEGK---------
MLBr_00238|M.leprae_Br4923 SVRLAPGTVLPPPTGVFPRYQPSEIEGADPVKSSSKRREHNKRRE
MMAR_4481|M.marinum_M GARLVPGTAMPSPTGVFPRYQP-------PPSETG----------
MUL_4654|M.ulcerans_Agy99 GARLVPGTAMPSPTGVFPRYQP-------PPSETG----------
MAV_1145|M.avium_104 GARLAPGTVLPPPTGVFPRYQAE------SEKVD-----------
*: **. :*.* *:**:**