For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TVEHGSSLDDVETTDRTESPSAGSARAGRLATIKLWAGRCAARWRSIVATALVAGAVGVTVSLYFVLYRP DQRVGDAAAHRAVQAASDGAAAVLSYAPESLNRDFAKAQSYLTGDFLAYYTKFTEQIAALAQQKQVTETA KVVRAAVSELHPDSAVVLVFINQTATSKQKPEPQTTASSARVTLTKVKGSWLISKLDPLS*
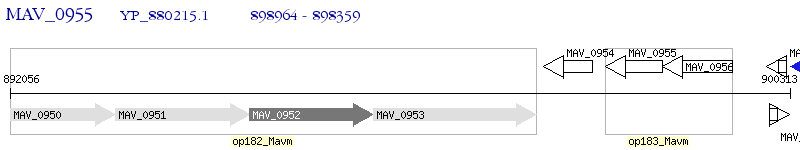
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_0955 | - | - | 100% (201) | hypothetical protein MAV_0955 |
| M. avium 104 | MAV_0937 | - | 6e-39 | 51.95% (154) | hypothetical protein MAV_0937 |
| M. avium 104 | MAV_1970 | - | 2e-19 | 34.52% (197) | hypothetical protein MAV_1970 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1397c | - | 2e-43 | 47.98% (198) | hypothetical protein Mb1397c |
| M. gilvum PYR-GCK | Mflv_2527 | - | 3e-39 | 45.92% (196) | hypothetical protein Mflv_2527 |
| M. tuberculosis H37Rv | Rv1362c | - | 3e-44 | 48.48% (198) | hypothetical protein Rv1362c |
| M. leprae Br4923 | MLBr_02598 | - | 7e-12 | 26.16% (172) | putative secreted protein |
| M. abscessus ATCC 19977 | MAB_4593c | - | 2e-13 | 27.04% (159) | hypothetical protein MAB_4593c |
| M. marinum M | MMAR_4703 | - | 3e-47 | 53.53% (170) | hypothetical protein MMAR_4703 |
| M. smegmatis MC2 155 | MSMEG_2865 | - | 1e-41 | 45.36% (194) | hypothetical protein MSMEG_2865 |
| M. thermoresistible (build 8) | TH_3191 | - | 3e-38 | 49.42% (172) | PUTATIVE twin-arginine translocation pathway signal |
| M. ulcerans Agy99 | MUL_3859 | - | 3e-41 | 46.35% (192) | hypothetical protein MUL_3859 |
| M. vanbaalenii PYR-1 | Mvan_4131 | - | 4e-38 | 53.25% (154) | twin-arginine translocation pathway signal |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_0955|M.avium_104 MTVEHGSSLDDVETTDRTESPSAG------------SARAGRLAT-----
MMAR_4703|M.marinum_M -------------------------------------MKVAGAAA-----
MSMEG_2865|M.smegmatis_MC2_155 MTVDEMTTTSDVEAD----------------------VEAGAEAG-----
Mb1397c|M.bovis_AF2122/97 -MTDDVRDVNTETTDATEVAEIDS------------AAGEAGDSATEAFD
Rv1362c|M.tuberculosis_H37Rv -MTDDVRDVNTETTDATEVAEIDS------------AAGEAGDSATEAFD
MUL_3859|M.ulcerans_Agy99 -MTDDARNLDSAETTQFSAADETA------------AIDPDALELEDPDD
Mflv_2527|M.gilvum_PYR-GCK -MSDREQTPDE-DVDATVVAEDPTPE----------DEVVTGTEP-----
Mvan_4131|M.vanbaalenii_PYR-1 -MTDENRTEDAAETDAALVEGDVIADVDSTDEADDVDEAAAGREPGD---
TH_3191|M.thermoresistible__bu -VTDEVKHTADTTEKAGGVAVGVA-D----------EAVAAAPDT-----
MAB_4593c|M.abscessus_ATCC_199 -MADDAADKDAKNADESAVTSDAEKAESSAPSSAAVESTAEGTTEAATAE
MLBr_02598|M.leprae_Br4923 ------------------------------------MAAAEGGGS-----
MAV_0955|M.avium_104 -------------------------------IKLWAGRCAARWRSIVATA
MMAR_4703|M.marinum_M -------------------------------VKRWTRRCLRRWRAILLTT
MSMEG_2865|M.smegmatis_MC2_155 -------------------------------TDSRLRRG-AHWRAIVVVG
Mb1397c|M.bovis_AF2122/97 TDSATESTAQKGQ----------------RHRDLWRMQVTLKPVPVILIL
Rv1362c|M.tuberculosis_H37Rv TDSATESTAQKGQ----------------RHRDLWRMQVTLKPVPVILIL
MUL_3859|M.ulcerans_Agy99 TGEASDADSPEAS----------------GKRDLWRIKVNLRPVSIILVV
Mflv_2527|M.gilvum_PYR-GCK -------EPAPAT----------------PRATSRRSGRVLIGAIVAAVL
Mvan_4131|M.vanbaalenii_PYR-1 -DDKAEADSGAAT----------------TKPPTRRPVRGLVTAILLAVA
TH_3191|M.thermoresistible__bu --------PGDNG----------------RDRTAWRAVLKEPSGLILLIV
MAB_4593c|M.abscessus_ATCC_199 VADFDAEDSEKTKDSGAVEDSDDEDISLEPDEEENPKRIRGRHWALTAAA
MLBr_02598|M.leprae_Br4923 --------------------------------WRNRRAGQRTAIAVVVAA
:
MAV_0955|M.avium_104 LVAGAVGVTVSLYFVLYRPDQRVGDAAAHRAVQAASDGAAAVLSYAPESL
MMAR_4703|M.marinum_M SLVAAVAFGAGYFYFVYRVDLQTDDGAAQQVVAAASDGAVALLSYSPETL
MSMEG_2865|M.smegmatis_MC2_155 LLVSSAALVTVLYLTQYRIDRQTDTEAVDEVVAAASSGTVALLSYAPDTL
Mb1397c|M.bovis_AF2122/97 LMLISGGATGWLYLEQYRPDQQTDSGAARAAGAAASDGTIALLSYSPDTL
Rv1362c|M.tuberculosis_H37Rv LMLISGGATGWLYLEQYRPDQQTDSGAARAAVAAASDGTIALLSYSPDTL
MUL_3859|M.ulcerans_Agy99 LVMISGGIAAWLYVKTYRPDQQVDPAVARQAVSAASDGTVALLSYSPDTL
Mflv_2527|M.gilvum_PYR-GCK MLAGSAGLAGWLYYNEYRPDRETDPAAAQVALEAAKTGTIALLSYSPESL
Mvan_4131|M.vanbaalenii_PYR-1 LLA-SAGMAGWLYLNDYRPDQQTDPAAAQVALDAARTGTVALLSYSPESL
TH_3191|M.thermoresistible__bu ALVAAAAVTASLWKFQYRLDVQTNTDAAQVAVDAATEGTIALLSYAPETL
MAB_4593c|M.abscessus_ATCC_199 IVAIAIVALAAIIGRLYE-EQRIEEQSSREALAAAQEFANVMINVDSAKL
MLBr_02598|M.leprae_Br4923 VLFVGSAAFAGAAVQPYLADRAT-VAVKLEVARTAANAITVLWTYTPENM
: . * : . :* .: . . .:
MAV_0955|M.avium_104 NRDFAKAQSYLTGDFLAYYTKFTEQIAALAQQK-QVTETAKVVRAAVSEL
MMAR_4703|M.marinum_M DHDFANARSRLTDNYLTYYRQFADQVVGPAAQRGRVTTTASVVRAAVAEL
MSMEG_2865|M.smegmatis_MC2_155 EADLAGARSKMTGDFLTYYGKFTSDVVAPAVQQKGITATAQIVRAGMMEI
Mb1397c|M.bovis_AF2122/97 DQDFATARSHLAGDFLSYYDQFTQQIVAPAAKQKSLKTTAKVVRAAVSEL
Rv1362c|M.tuberculosis_H37Rv DQDFATARSHLAGDFLSYYDQFTQQIVAPAAKQKSLKTTAKVVRAAVSEL
MUL_3859|M.ulcerans_Agy99 DDDFAAAKSHLSGDFLAYYEQFTEQFVAPAAKQKALKTSAQVVRAAVSEL
Mflv_2527|M.gilvum_PYR-GCK DQDFANAKSKLTGDFLDYYTQFTEQIVTPAAKEKSVKTAASVVRAGVSQI
Mvan_4131|M.vanbaalenii_PYR-1 DQDFANAKAKLTGEFLSYYTQFTEQIVTPAAKEKSVKTAASVVRAAVADI
TH_3191|M.thermoresistible__bu QEDFAAAKEHLTGEFLDYYTEFTDTVVTPAAMEKQVSTSANVVKAAISEI
MAB_4593c|M.abscessus_ATCC_199 DESSNKTLERSTGDFKQKYSE-SSTTLRQVMIENKAKATGVVVDSAVKSA
MLBr_02598|M.leprae_Br4923 DTLADRAATYVSGDFGAQYRKFVDAIVGP-NKQAKITNSTEVTGVAVESL
: : :.:: * : . . . : :. .: .
MAV_0955|M.avium_104 HPDSAVVLVFINQTATS-KQKPEPQTTASSARVTLTKVKGSWLISKLDPL
MMAR_4703|M.marinum_M HPNSAVVLVFVKQKTAS-KQKPEPIVTSSSIRVTLTKVNSSWLIERFDPM
MSMEG_2865|M.smegmatis_MC2_155 RPDSAKVLVFLNQETSS-KERPDPSLTASSVVVSLTKVDGNWLISAFDPV
Mb1397c|M.bovis_AF2122/97 HPDSAVVLVFVDQSTTS-KDSPNPSMAASSVMVTLAKVDGNWLITKFTPV
Rv1362c|M.tuberculosis_H37Rv HPDSAVVLVFVDQSTTS-KDSPNPSMAASSVMVTLAKVDGNWLITKFTPV
MUL_3859|M.ulcerans_Agy99 HPGSAEVLLFVDQSTTS-KDHPDPTMMPISVLVSMAQIDGKWLITKFTPV
Mflv_2527|M.gilvum_PYR-GCK AADSAEVLVFINQTTTS-KENPDGAFAASSVKVGMKKIDGNWLIASFDPV
Mvan_4131|M.vanbaalenii_PYR-1 QPESAEVLVFINQTTTS-KENPDGAFAASSVKVGLKKIDGNWLIESFDPV
TH_3191|M.thermoresistible__bu HPDSAVVLVFLNQMTTS-KENPDGAFAASSVKVGLKKVDGRWLIEAFDPV
MAB_4593c|M.abscessus_ATCC_199 APDKVEVLLFIDQAVSN-VAIPDARMDRSRVQITMQKVDGRWLASDVELT
MLBr_02598|M.leprae_Br4923 DASNAIAIVYTNTTSTSPLTKNIPALKYLSYRLFMKRSAVRWLVTRMTTI
. .. .::: . :. : : : ** .
MAV_0955|M.avium_104 S--------
MMAR_4703|M.marinum_M ---------
MSMEG_2865|M.smegmatis_MC2_155 ---------
Mb1397c|M.bovis_AF2122/97 ---------
Rv1362c|M.tuberculosis_H37Rv ---------
MUL_3859|M.ulcerans_Agy99 ---------
Mflv_2527|M.gilvum_PYR-GCK ---------
Mvan_4131|M.vanbaalenii_PYR-1 ---------
TH_3191|M.thermoresistible__bu ---------
MAB_4593c|M.abscessus_ATCC_199 ---------
MLBr_02598|M.leprae_Br4923 TSLDLTPQL