For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SEDNRSSDAADDSADRPEPGRPRARRNVKVVPIVLISLLLISGAGTAWLYFKEYLPDKRTDARVASAVVN AASDGTVALLSYSPDTLDKDFAAAKSHLSGDFLSYYNQFTEQIVAPAAKQKSLKTNARVLGAAVSDLRPD SAVVLVLVDQSTTSKDNPDPAMASSSVLVSLTKVNGKWLITKFDPV*
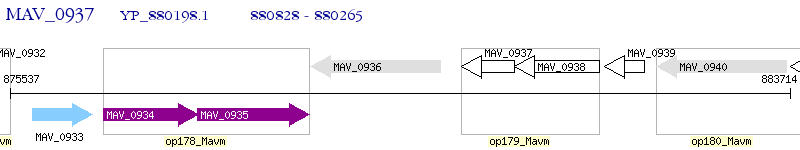
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_0937 | - | - | 100% (187) | hypothetical protein MAV_0937 |
| M. avium 104 | MAV_0955 | - | 5e-39 | 51.95% (154) | hypothetical protein MAV_0955 |
| M. avium 104 | MAV_2071 | - | 9e-26 | 36.84% (190) | hypothetical protein MAV_2071 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1397c | - | 2e-62 | 64.48% (183) | hypothetical protein Mb1397c |
| M. gilvum PYR-GCK | Mflv_2527 | - | 5e-49 | 57.14% (175) | hypothetical protein Mflv_2527 |
| M. tuberculosis H37Rv | Rv1362c | - | 3e-63 | 65.03% (183) | hypothetical protein Rv1362c |
| M. leprae Br4923 | MLBr_02598 | - | 1e-13 | 27.39% (157) | putative secreted protein |
| M. abscessus ATCC 19977 | MAB_4593c | - | 5e-10 | 24.48% (192) | hypothetical protein MAB_4593c |
| M. marinum M | MMAR_3995 | - | 6e-59 | 62.96% (189) | hypothetical protein MMAR_3995 |
| M. smegmatis MC2 155 | MSMEG_4770 | - | 5e-47 | 47.50% (200) | hypothetical protein MSMEG_4770 |
| M. thermoresistible (build 8) | TH_3191 | - | 1e-43 | 50.54% (186) | PUTATIVE twin-arginine translocation pathway signal |
| M. ulcerans Agy99 | MUL_3859 | - | 1e-59 | 63.49% (189) | hypothetical protein MUL_3859 |
| M. vanbaalenii PYR-1 | Mvan_4131 | - | 3e-50 | 52.04% (196) | twin-arginine translocation pathway signal |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_3995|M.marinum_M -MTDDVRNLDSAETTQFSAADETAAI--------DPDALELEDPDDTGEA
MUL_3859|M.ulcerans_Agy99 -MTDDARNLDSAETTQFSAADETAAI--------DPDALELEDPDDTGEA
MAV_0937|M.avium_104 -MSEDNRSSD--------AADDS---------------------------
Mb1397c|M.bovis_AF2122/97 -MTDDVRDVNTETTDATEVAEIDSAA--------GEAGDSATEAFDTDSA
Rv1362c|M.tuberculosis_H37Rv -MTDDVRDVNTETTDATEVAEIDSAA--------GEAGDSATEAFDTDSA
Mflv_2527|M.gilvum_PYR-GCK -MSDREQTPDE-DVDATVVAEDPTPE----------DEVVTGTEP-----
Mvan_4131|M.vanbaalenii_PYR-1 -MTDENRTEDAAETDAALVEGDVIADVDSTDEADDVDEAAAGREPGDDDK
MSMEG_4770|M.smegmatis_MC2_155 -MSAQDQTSDAVEAPETV------------------DQAVTATEP-----
TH_3191|M.thermoresistible__bu VTDEVKHTADTTEKAGGVAVG-------------VADEAVAAAPD-----
MAB_4593c|M.abscessus_ATCC_199 -MADDAADKDAKNADESAVTSDAEKAESSAPSSAAVESTAEGTTEAATAE
MLBr_02598|M.leprae_Br4923 --------------------------------------------------
MMAR_3995|M.marinum_M SDADSPEASGKRDLWRIKVNLRPV-------------------SIILVVL
MUL_3859|M.ulcerans_Agy99 SDADSPEASGKRDLWRIKVNLRPV-------------------SIILVVL
MAV_0937|M.avium_104 --ADRPEP-GRP---RARRNVKVV-------------------PIVLISL
Mb1397c|M.bovis_AF2122/97 TESTAQKGQRHRDLWRMQVTLKPV-------------------PVILILL
Rv1362c|M.tuberculosis_H37Rv TESTAQKGQRHRDLWRMQVTLKPV-------------------PVILILL
Mflv_2527|M.gilvum_PYR-GCK ---EPAPATPRATSRRSGRVLIGA-------------------IVAAVLM
Mvan_4131|M.vanbaalenii_PYR-1 AEADSGAATTKPPTRRPVRGLVTA-------------------ILLAVAL
MSMEG_4770|M.smegmatis_MC2_155 ---APAHAPSRG-VLKAIRAKTVP-------------------IMLGIAL
TH_3191|M.thermoresistible__bu ---TPGDNGRDRTAWRAVLKEPSG-------------------LILLIVA
MAB_4593c|M.abscessus_ATCC_199 VADFDAEDSEKTKDSGAVEDSDDEDISLEPDEEENPKRIRGRHWALTAAA
MLBr_02598|M.leprae_Br4923 --MAAAEGGGSWRNRRAGQRTAIA--------------------VVVAAV
MMAR_3995|M.marinum_M VMISGGIAAWLYVKTYRPDQQVDPAVARQAVSAASDGTVALLSYSPDTLD
MUL_3859|M.ulcerans_Agy99 VMISGGIAAWLYVKTYRPDQQVDPAVARQAVSAASDGTVALLSYSPDTLD
MAV_0937|M.avium_104 LLISGAGTAWLYFKEYLPDKRTDARVASAVVNAASDGTVALLSYSPDTLD
Mb1397c|M.bovis_AF2122/97 MLISGGATGWLYLEQYRPDQQTDSGAARAAGAAASDGTIALLSYSPDTLD
Rv1362c|M.tuberculosis_H37Rv MLISGGATGWLYLEQYRPDQQTDSGAARAAVAAASDGTIALLSYSPDTLD
Mflv_2527|M.gilvum_PYR-GCK LAGSAGLAGWLYYNEYRPDRETDPAAAQVALEAAKTGTIALLSYSPESLD
Mvan_4131|M.vanbaalenii_PYR-1 LA-SAGMAGWLYLNDYRPDQQTDPAAAQVALDAARTGTVALLSYSPESLD
MSMEG_4770|M.smegmatis_MC2_155 VA-SAALATFVYLTQYRVDKETDPAAAEVALKAATDGTVALLSYAPESLD
TH_3191|M.thermoresistible__bu LVAAAAVTASLWKFQYRLDVQTNTDAAQVAVDAATEGTIALLSYAPETLQ
MAB_4593c|M.abscessus_ATCC_199 IVAIAIVALAAIIGRLYEEQRIEEQSSREALAAAQEFANVMINVDSAKLD
MLBr_02598|M.leprae_Br4923 LFVGSAAFAGAAVQPYLADRATVAVKLEVARTAAN-AITVLWTYTPENMD
: . : . ** .: . . .::
MMAR_3995|M.marinum_M DDFAAAKSHLSGDFLAYYEQFTEQFVAPAAKQKALKTSAQVVRAAVSELH
MUL_3859|M.ulcerans_Agy99 DDFAAAKSHLSGDFLAYYEQFTEQFVAPAAKQKALKTSAQVVRAAVSELH
MAV_0937|M.avium_104 KDFAAAKSHLSGDFLSYYNQFTEQIVAPAAKQKSLKTNARVLGAAVSDLR
Mb1397c|M.bovis_AF2122/97 QDFATARSHLAGDFLSYYDQFTQQIVAPAAKQKSLKTTAKVVRAAVSELH
Rv1362c|M.tuberculosis_H37Rv QDFATARSHLAGDFLSYYDQFTQQIVAPAAKQKSLKTTAKVVRAAVSELH
Mflv_2527|M.gilvum_PYR-GCK QDFANAKSKLTGDFLDYYTQFTEQIVTPAAKEKSVKTAASVVRAGVSQIA
Mvan_4131|M.vanbaalenii_PYR-1 QDFANAKAKLTGEFLSYYTQFTEQIVTPAAKEKSVKTAASVVRAAVADIQ
MSMEG_4770|M.smegmatis_MC2_155 QDFATAKTHLTGDFLSYYTDFTTKVVAPAAQQKKVKTKATVVNGAVSEIK
TH_3191|M.thermoresistible__bu EDFAAAKEHLTGEFLDYYTEFTDTVVTPAAMEKQVSTSANVVKAAISEIH
MAB_4593c|M.abscessus_ATCC_199 ESSNKTLERSTGDFKQKYSE-SSTTLRQVMIENKAKATGVVVDSAVKSAA
MLBr_02598|M.leprae_Br4923 TLADRAATYVSGDFGAQYRKFVDAIVGPNK-QAKITNSTEVTGVAVESLD
: :*:* * . : : . * .: .
MMAR_3995|M.marinum_M PGSAEVLLFVDQSTSS-KDHPDPTMMPISVLVSMAQIDGKWLITKFTPV-
MUL_3859|M.ulcerans_Agy99 PGSAEVLLFVDQSTTS-KDHPDPTMMPISVLVSMAQIDGKWLITKFTPV-
MAV_0937|M.avium_104 PDSAVVLVLVDQSTTS-KDNPDPAMASSSVLVSLTKVNGKWLITKFDPV-
Mb1397c|M.bovis_AF2122/97 PDSAVVLVFVDQSTTS-KDSPNPSMAASSVMVTLAKVDGNWLITKFTPV-
Rv1362c|M.tuberculosis_H37Rv PDSAVVLVFVDQSTTS-KDSPNPSMAASSVMVTLAKVDGNWLITKFTPV-
Mflv_2527|M.gilvum_PYR-GCK ADSAEVLVFINQTTTS-KENPDGAFAASSVKVGMKKIDGNWLIASFDPV-
Mvan_4131|M.vanbaalenii_PYR-1 PESAEVLVFINQTTTS-KENPDGAFAASSVKVGLKKIDGNWLIESFDPV-
MSMEG_4770|M.smegmatis_MC2_155 PESAVVLLFINQTTTS-QEIPDGSFATSSVKVGLTKIDGNWLISTFDPV-
TH_3191|M.thermoresistible__bu PDSAVVLVFLNQMTTS-KENPDGAFAASSVKVGLKKVDGRWLIEAFDPV-
MAB_4593c|M.abscessus_ATCC_199 PDKVEVLLFIDQAVSN-VAIPDARMDRSRVQITMQKVDGRWLASDVELT-
MLBr_02598|M.leprae_Br4923 ASNAIAIVYTNTTSTSPLTKNIPALKYLSYRLFMKRSAVRWLVTRMTTIT
. .. .:: : :. : : : : .** .
MMAR_3995|M.marinum_M --------
MUL_3859|M.ulcerans_Agy99 --------
MAV_0937|M.avium_104 --------
Mb1397c|M.bovis_AF2122/97 --------
Rv1362c|M.tuberculosis_H37Rv --------
Mflv_2527|M.gilvum_PYR-GCK --------
Mvan_4131|M.vanbaalenii_PYR-1 --------
MSMEG_4770|M.smegmatis_MC2_155 --------
TH_3191|M.thermoresistible__bu --------
MAB_4593c|M.abscessus_ATCC_199 --------
MLBr_02598|M.leprae_Br4923 SLDLTPQL