For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MLDGKVVVISGVGPGLGTTLAHRCADSGADLVLAARTAERLEKVAKQVNDGGHRALAVRTDITDDDEVAY LVETTMATYGRADVLINNAFRVPSMKPLAGTSFQHIRDAIELSALGALRLIQAFTPALETAHGSIVNVNS MVLRHSQAKYGAYKMAKSALLSMSQSLATELGEKGIRVNSVAPGYIWGDTLQAYFEHQAGKYGTTVEQIY AATAANSDLKRLPTEDEVASAIMFLASDLSSGITGQTLDVNCGEYHT
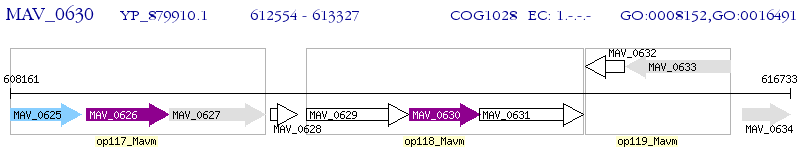
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_0630 | - | - | 100% (257) | short chain dehydrogenase |
| M. avium 104 | MAV_1391 | - | 5e-33 | 35.83% (254) | short-chain dehydrogenase/reductase SDR |
| M. avium 104 | MAV_1046 | - | 3e-29 | 33.20% (256) | short chain dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3560c | - | 1e-122 | 83.27% (257) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_1530 | - | 1e-110 | 75.69% (255) | short chain dehydrogenase |
| M. tuberculosis H37Rv | Rv3530c | - | 1e-122 | 83.27% (257) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_02565 | fabG | 4e-15 | 28.52% (263) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. abscessus ATCC 19977 | MAB_4178c | - | 4e-95 | 65.75% (254) | short chain dehydrogenase |
| M. marinum M | MMAR_5018 | - | 1e-116 | 80.16% (257) | short-chain alcohol dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_5931 | - | 1e-113 | 79.22% (255) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_3268 | - | 1e-110 | 79.13% (254) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_4092 | - | 1e-107 | 78.46% (246) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_5228 | - | 1e-108 | 75.29% (255) | short chain dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1530|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5228|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5931|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_5018|M.marinum_M --------------------------------------------------
MUL_4092|M.ulcerans_Agy99 --------------------------------------------------
Mb3560c|M.bovis_AF2122/97 --------------------------------------------------
Rv3530c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_0630|M.avium_104 --------------------------------------------------
TH_3268|M.thermoresistible__bu --------------------------------------------------
MAB_4178c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_02565|M.leprae_Br4923 MAPKCSSDLFSQIVSSGPGSFVAKQLGVPRPETLRRYRPSDAPLCGSLLI
Mflv_1530|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5228|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5931|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_5018|M.marinum_M --------------------------------------------------
MUL_4092|M.ulcerans_Agy99 --------------------------------------------------
Mb3560c|M.bovis_AF2122/97 --------------------------------------------------
Rv3530c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_0630|M.avium_104 --------------------------------------------------
TH_3268|M.thermoresistible__bu --------------------------------------------------
MAB_4178c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_02565|M.leprae_Br4923 GGDGRLVEPLRATLEKDYDLVSNNLGGRWTDSFGGLVFDATGITAPAELK
Mflv_1530|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5228|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5931|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_5018|M.marinum_M --------------------------------------------------
MUL_4092|M.ulcerans_Agy99 --------------------------------------------------
Mb3560c|M.bovis_AF2122/97 --------------------------------------------------
Rv3530c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_0630|M.avium_104 --------------------------------------------------
TH_3268|M.thermoresistible__bu --------------------------------------------------
MAB_4178c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_02565|M.leprae_Br4923 GLHNFFTPLLRNLGRCARVVVIGTTPDATTSTDERIAQRALEGFSRSLGK
Mflv_1530|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5228|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5931|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_5018|M.marinum_M --------------------------------------------------
MUL_4092|M.ulcerans_Agy99 --------------------------------------------------
Mb3560c|M.bovis_AF2122/97 --------------------------------------------------
Rv3530c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_0630|M.avium_104 --------------------------------------------------
TH_3268|M.thermoresistible__bu --------------------------------------------------
MAB_4178c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_02565|M.leprae_Br4923 ELRRGATVALVYLSPDAKPAATGLESTMRFILSAKSAYVNGQVFYVGAAD
Mflv_1530|M.gilvum_PYR-GCK ---MSEALPLLENKVVVISGVGPALGTTLARRCAEAGADLVLAART--VE
Mvan_5228|M.vanbaalenii_PYR-1 -----MAFGLLRDKVVVISGVGPALGTTLARRCAEEGADLVLAART--VE
MSMEG_5931|M.smegmatis_MC2_155 ------MGGLLTDKVVVISGVGPALGTTLARRCAEQGADLVLAART--VE
MMAR_5018|M.marinum_M ------MPGLLDGKTLVISGVGPGLGTTLAHRCAAEGADLVLAARS--GD
MUL_4092|M.ulcerans_Agy99 --------------------MGPGLGTTLAHRCAAEGADLVLAARI--GD
Mb3560c|M.bovis_AF2122/97 ------MTGMLKRKVIVVSGVGPGLGTTLAHRCARDGADLVLAARS--AE
Rv3530c|M.tuberculosis_H37Rv ------MTGMLKRKVIVVSGVGPGLGTTLAHRCARDGADLVLAARS--AE
MAV_0630|M.avium_104 ---------MLDGKVVVISGVGPGLGTTLAHRCADSGADLVLAART--AE
TH_3268|M.thermoresistible__bu -----MTMLSLDNKVVVISGVGPALGTTLARRCAAAGADLVLAART--AE
MAB_4178c|M.abscessus_ATCC_199 -----MSLLNFDGKVVVVSGIGPGLGAALAIKFAEAGADVVLAART--QS
MLBr_02565|M.leprae_Br4923 STPPTNWDRPLENKVAIVTGAARGIGAAIAEVVARDGARVVAIDVESAAE
. .:*:::* * ** :* .
Mflv_1530|M.gilvum_PYR-GCK RLEDVATQITDLGRRAVSVGTDITDEAQVANLVEESLKAYG-KVDVLINN
Mvan_5228|M.vanbaalenii_PYR-1 RLDDVARQITDLGRRAVSVGTDITDEAQVNNLVDESLKAYG-KVDVLINN
MSMEG_5931|M.smegmatis_MC2_155 RLEDVAKQVTDTGRRALSVGTDITDDAQVAHLVDETMKAYG-RVDVVINN
MMAR_5018|M.marinum_M RLDELAKQIIDKGRQAIAVRADITDDDQVSNLVDATLEAYG-KIDVLVNN
MUL_4092|M.ulcerans_Agy99 RLDELAKQIIDKGRQAIAVRADITDDDQVSNLVDATLEAYG-KIDVLVNN
Mb3560c|M.bovis_AF2122/97 RLDDVAKQIIDTGRRAVAVRTDITDDDDVSNLVQATLAAYG-KADVLINN
Rv3530c|M.tuberculosis_H37Rv RLDDVAKQIIDTGRRAVAVRTDITDDDDVSNLVQATLAAYG-KADVLINN
MAV_0630|M.avium_104 RLEKVAKQVNDGGHRALAVRTDITDDDEVAYLVETTMATYG-RADVLINN
TH_3268|M.thermoresistible__bu RLEEVAGQITATGRRAVAVPTDITDEAQVANLVAETNKAYG-RVDVLINN
MAB_4178c|M.abscessus_ATCC_199 RLDEVARQIAATGRRALAVATDITDEASAANLVERATEEFG-GIDVLVNN
MLBr_02565|M.leprae_Br4923 ALDETANRVG-----GTTLSLDVTADDAVDKITEHLHDYQGGHADILVNN
*:. * :: . :: *:* : . :. * *:::**
Mflv_1530|M.gilvum_PYR-GCK AFRVPSMKPFANTTFEHMRDAIELTVFGALRMVQAFTPALAEANG-AVVN
Mvan_5228|M.vanbaalenii_PYR-1 AFRVPSMKPFADTTFEHMRDAIELTVFGALRMVQAFSPALAEANG-AVVN
MSMEG_5931|M.smegmatis_MC2_155 AFRVPSMKPFANTTFEHMRDAIELTVFGALRLIQGFTPALEESKG-AVVN
MMAR_5018|M.marinum_M AFRVPSMKPLAETTFQHIRDAIELSALGALRVIAGFTRALAETNG-SIVN
MUL_4092|M.ulcerans_Agy99 AFRVPSMKPLAETTFQHIRDAIELSALGALRVIAGFTRALAETNG-SIVN
Mb3560c|M.bovis_AF2122/97 AFRVPSMKPLAGTTFEHIRDAIELSALGTLRLIQAFTPALAQSHG-AIVN
Rv3530c|M.tuberculosis_H37Rv AFRVPSMKPLAGTTFEHIRDAIELSALGTLRLIQAFTPALAQSHG-AIVN
MAV_0630|M.avium_104 AFRVPSMKPLAGTSFQHIRDAIELSALGALRLIQAFTPALETAHG-SIVN
TH_3268|M.thermoresistible__bu AFRVPSMKPLANTGFDHIREAIELTVLGALRLTQALTPALAQANG-SVVN
MAB_4178c|M.abscessus_ATCC_199 AFSIPSMKSLEKTDYQHIRDSVELTVVGTLRLTQRVAAALAQSKG-AVVN
MLBr_02565|M.leprae_Br4923 AG-ITRDKLLANMDDCRWDAVLEVNLLAPQRLTEGLVGNGSISKGGRVIV
* :. * : : :*:. ... *: . ::* ::
Mflv_1530|M.gilvum_PYR-GCK VNSMVVRHSQAKYGAYKMAKSALLAMSQTLATELGDQGIRVNSVLPGYIW
Mvan_5228|M.vanbaalenii_PYR-1 VNSMVVRHSQAKYGAYKMAKSALLAMSQTLATELGEQGIRVNSVLPGYIW
MSMEG_5931|M.smegmatis_MC2_155 VNSMVVRHSQAKYGAYKMAKSALLAMSQTLATELGEKGIRVNSVLPGYIW
MMAR_5018|M.marinum_M VNSMVLRHSQPKFGAYKMAKSTLLAMSHSLAAELGVKGIRVNSVAPGYIW
MUL_4092|M.ulcerans_Agy99 VNSMVLRHSQPKFGADKTAKSTLLAMSHSLAAELGVKGIWVNSVAPGYIW
Mb3560c|M.bovis_AF2122/97 VNSMVIRHSQPKYGTYKMAKSVLLAMSHSLATELGEQGIRVNSVAPGYIW
Rv3530c|M.tuberculosis_H37Rv VNSMVIRHSQPKYGTYKMAKSVLLAMSHSLATELGEQGIRVNSVAPGYIW
MAV_0630|M.avium_104 VNSMVLRHSQAKYGAYKMAKSALLSMSQSLATELGEKGIRVNSVAPGYIW
TH_3268|M.thermoresistible__bu VNSMVIRHSQPKYGAYKMAKAALLAMSQSLASELGEQGIRVNSVVPGYIW
MAB_4178c|M.abscessus_ATCC_199 INSMVIRHSQVRYGSYKIAKAALLAMSQSLATELGPQGVRVNSVVPGYMW
MLBr_02565|M.leprae_Br4923 LSSMAGIAGNRGQTNYATAKAGVIGLTQALAPGLAEKGITINAVAPGFIE
:.**. .: **: ::.::::**. *. :*: :*:* **::
Mflv_1530|M.gilvum_PYR-GCK GGTLESYFSHQAGKYGTTVDEIYKATAAASDLKRLPTEDEVASAILFMAC
Mvan_5228|M.vanbaalenii_PYR-1 GGTLESYFTHQAGKYGTTVEEIYKATAAASDLKRLPTEDEVASAILFMAS
MSMEG_5931|M.smegmatis_MC2_155 GGTLKSYFEHQAGKYGTSVEDIYNAAAAGSDLKRLPTEDEVASAILFMAS
MMAR_5018|M.marinum_M GDTLQGYFEHQAGKYGTTVEQIYQATAAQSDLKRLPTEDEVASAILFMAS
MUL_4092|M.ulcerans_Agy99 GDTLQGYFEHQAGKYGTTVEKIYQATAAQSDLKRLPTEDEVASAVLFMAS
Mb3560c|M.bovis_AF2122/97 GDTLKSYFDHQAGKYGTTVDQIYQATAANSDLKRLPTEDEVASAILFLAS
Rv3530c|M.tuberculosis_H37Rv GDTLKSYFDHQAGKYGTTVDQIYQATAANSDLKRLPTEDEVASAILFLAS
MAV_0630|M.avium_104 GDTLQAYFEHQAGKYGTTVEQIYAATAANSDLKRLPTEDEVASAIMFLAS
TH_3268|M.thermoresistible__bu GETLKGYFQHQAGKYNTTVDQIYQATAAQSDLKRLPTEDEVASAILFFAS
MAB_4178c|M.abscessus_ATCC_199 GENLKGYFEHQAKKYGGTVEQIYEYTAANSDLKRLPTTDEVANAVLFLAS
MLBr_02565|M.leprae_Br4923 T-QMTAAIPLATREVGRRLNSLLQGGLP----------VDVAETIAYFAI
: . : : : . ::.: . :**.:: ::*
Mflv_1530|M.gilvum_PYR-GCK DLSSGITGQALDVNCGEFKA-
Mvan_5228|M.vanbaalenii_PYR-1 DLSSGITGQALDVNCGEFKA-
MSMEG_5931|M.smegmatis_MC2_155 DLASGITGQALDVNCGEYKA-
MMAR_5018|M.marinum_M DLSSGITGQTLDVNCGEYHT-
MUL_4092|M.ulcerans_Agy99 DLSSGITGQTLDVNCGEYHT-
Mb3560c|M.bovis_AF2122/97 DLASGITGQTLDVNCGEYHT-
Rv3530c|M.tuberculosis_H37Rv DLASGITGQTLDVNCGEYHT-
MAV_0630|M.avium_104 DLSSGITGQTLDVNCGEYHT-
TH_3268|M.thermoresistible__bu DLSSGITGQTLDVNCGEYKV-
MAB_4178c|M.abscessus_ATCC_199 DLASGITGQALDVNCGEFHA-
MLBr_02565|M.leprae_Br4923 PASNAVTGNVIRVCGQAMLGA
:..:**:.: *