For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TNFDDLTLLNDVLRAIYSADTADFPQVLDDLTAASARWVPGAQEAGITVTSRQNEVSTPSVTDDCARLLD EFQQRYLEGPCLHAAWTRKVVVVDDLRTDSRWPKYQADALARTPIRSILSLPMYAGELSMGALNFYAERP HAFSDDSRRMAALFATLGSLAWSNVVRTKQFKEALSTRDMIGQAKGILMERYELDDETAFNTLIKLSQSM NTPLRDIARRVIDDTTQR*
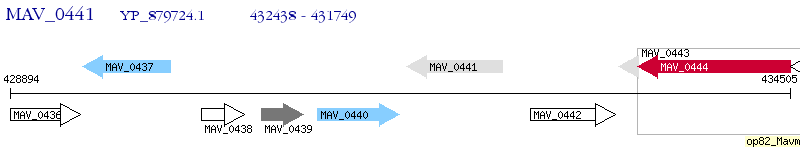
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_0441 | - | - | 100% (229) | ANTAR domain-containing protein |
| M. avium 104 | MAV_4102 | - | 2e-45 | 44.12% (204) | ANTAR domain-containing protein |
| M. avium 104 | MAV_5172 | - | 2e-35 | 40.10% (202) | F420-dependent glucose-6-phosphate dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3842 | - | 5e-51 | 45.28% (212) | response regulator receiver/ANTAR domain-containing protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2534c | - | 7e-37 | 38.07% (197) | hypothetical protein MAB_2534c |
| M. marinum M | MMAR_1808 | - | 3e-23 | 29.13% (230) | hypothetical protein MMAR_1808 |
| M. smegmatis MC2 155 | MSMEG_5554 | - | 2e-51 | 44.39% (214) | ANTAR domain-containing protein |
| M. thermoresistible (build 8) | TH_2526 | - | 3e-21 | 30.22% (225) | PUTATIVE putative response regulator receiver and ANTAR domain |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_2553 | - | 1e-52 | 46.26% (214) | response regulator receiver/ANTAR domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3842|M.gilvum_PYR-GCK MRAPRARGSGNLDVLLTRNTRTVTVNDSADVEAAHLRVAELVQGLHNRPD
Mvan_2553|M.vanbaalenii_PYR-1 ------------------------MSDPAGVEVTHLRVAELVQGLHNRPD
MSMEG_5554|M.smegmatis_MC2_155 -------------------------MSVSQNEAISQQISNLIRDVHTRRA
MAV_0441|M.avium_104 ---------------------------MTNFDDLTL-LNDVLRAIYSADT
MAB_2534c|M.abscessus_ATCC_199 ------------------------MTDEAAAGAFSAKIADAARRMQQEHS
TH_2526|M.thermoresistible__bu -------------------------------MGLNATVEKLIRKAR-ASG
MMAR_1808|M.marinum_M ---------------MTATPKDSNPDSVPGGTTADPATVFAALAEIIYQG
Mflv_3842|M.gilvum_PYR-GCK ADSDTVVAELAEHAAAEIPGAQYAGITLTRNAKHVETPAATHHWPLLLDK
Mvan_2553|M.vanbaalenii_PYR-1 TDSDTVVAELAELAAAEIEGAQYAGITLTRNAKQIETPAATHHWPLLLDK
MSMEG_5554|M.smegmatis_MC2_155 TDVDAVLGELTESAVEYVAGAQYAGITIAGRDGKVRTAAATGEYPEILDE
MAV_0441|M.avium_104 ADFPQVLDDLTAASARWVPGAQEAGITVTSRQNEVSTPSVTDDCARLLDE
MAB_2534c|M.abscessus_ATCC_199 S-VEMTLRSITTAALETVPGVEQAGITLVTGRYEIESRAATSETPRKLDD
TH_2526|M.thermoresistible__bu ADTCAVLTELTRSAVGDWPSIQHAGITVVEDGDVVRSIAATDGHVLVVDN
MMAR_1808|M.marinum_M AQPAEMYSAICIAATLIVPGCDHASLLVRTN-DRYITVGASDRLAQHVDE
: : : . : *.: : : ..: :*.
Mflv_3842|M.gilvum_PYR-GCK IQQRYLEGPCLTAAWEEQTVHVRDLQTEERFPNYRRDALAETPIRSIMAF
Mvan_2553|M.vanbaalenii_PYR-1 IQQRYLEGPCLTAAWEEKTVHVRNLETDERFPNYRRDALAETPIRSIMAF
MSMEG_5554|M.smegmatis_MC2_155 IQQRVENGPCLAAAWEQHVIRIDDMESEQRWPAYCRQALLETPIRSVVAF
MAV_0441|M.avium_104 FQQRYLEGPCLHAAWTRKVVVVDDLRTDSRWPKYQADALARTPIRSILSL
MAB_2534c|M.abscessus_ATCC_199 IQQELREGPCVQSAWEHETVYAEDLAQTTPWPRFA-EAAVKLGVRSMLSF
TH_2526|M.thermoresistible__bu IARRLSTGPGIDAALDEDSYRIDDLRADSRWPLFADKVAASTPIRSVIAI
MMAR_1808|M.marinum_M LERKAGDGPCIDAIEEETPQIETDLTTPTQWPKFAAALLAQTPVRGAMGF
: :. ** : : . :: :* : :*. :.:
Mflv_3842|M.gilvum_PYR-GCK QL------------FIADESLGALNVYSEEPDVFGSEARTLGLIFAAHSS
Mvan_2553|M.vanbaalenii_PYR-1 QL------------FIAGETLGALNVYSEQPEAFGDESRTLGLIFAAHSS
MSMEG_5554|M.smegmatis_MC2_155 QL------------YADNQTMGALNFYSETAGAFDAEAVESGLIIATHAA
MAV_0441|M.avium_104 PM------------YAGELSMGALNFYAERPHAFSDDSRRMAALFATLGS
MAB_2534c|M.abscessus_ATCC_199 QL------------FTFGENLGALNLYAQSPGAFGADSYDAGLALATHAA
TH_2526|M.thermoresistible__bu PVGNPIRNPIRNPIASAGTVRAALSLYSDLPGAFDDEVEAVAATYAEHIG
MMAR_1808|M.marinum_M RL------------LVDKRKGAALNLFSETPNSFDTESAGRAAVLASFAS
: .**..::: . *. : . * .
Mflv_3842|M.gilvum_PYR-GCK VAWNSARR--------EAQFRRALESRDVIGQAKGMIMERYRVDAVRAFE
Mvan_2553|M.vanbaalenii_PYR-1 VAWNSARR--------EEQFRQALSSRDVIGQAKGMIMERYRVDAVQAFE
MSMEG_5554|M.smegmatis_MC2_155 LMWSLMRR--------DEQFRSALASRDLIGQAKGMLMERYRIDAGQAFE
MAV_0441|M.avium_104 LAWSNVVR--------TKQFKEALSTRDMIGQAKGILMERYELDDETAFN
MAB_2534c|M.abscessus_ATCC_199 IVLVAAQR--------ESQWASALASRDVIGQAKGMLMERFSINAAEAFT
TH_2526|M.thermoresistible__bu RVLTARRGRRRAPVSGSARAQGSVRGQDAIETSKAQLMKHFGVDPAAALT
MMAR_1808|M.marinum_M VAVNAIAHG-----EDAASLRRGLLSNREIGKAVGMLMLLNGFTEEEAFD
.: . * : . :* . *:
Mflv_3842|M.gilvum_PYR-GCK LLRKLSQDSNVPLSKVAADLVAQAQTSNGQTG------------------
Mvan_2553|M.vanbaalenii_PYR-1 LLRKLSQDSNVPLIKIATSLVSDAQASAD---------------------
MSMEG_5554|M.smegmatis_MC2_155 VLKKLSQGANTPLVDVAREIVTTAERPADVSTQ-----------------
MAV_0441|M.avium_104 TLIKLSQSMNTPLRDIARRVIDDTTQR-----------------------
MAB_2534c|M.abscessus_ATCC_199 VIRTLSQDFNTKLVEIARAIVEDRPLG-----------------------
TH_2526|M.thermoresistible__bu VLLRLSRECNRSLEDVARLLVGPRPVAEPPSQATATATPDHAGGRPPVWA
MMAR_1808|M.marinum_M LLRHHSQSLNIKLADVARTVIENRGQLPPEALETDDTVNPTVSG------
: *: * * .:* ::
Mflv_3842|M.gilvum_PYR-GCK ----
Mvan_2553|M.vanbaalenii_PYR-1 ----
MSMEG_5554|M.smegmatis_MC2_155 ----
MAV_0441|M.avium_104 ----
MAB_2534c|M.abscessus_ATCC_199 ----
TH_2526|M.thermoresistible__bu EGAA
MMAR_1808|M.marinum_M ----