For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SVSQNEAISQQISNLIRDVHTRRATDVDAVLGELTESAVEYVAGAQYAGITIAGRDGKVRTAAATGEYPE ILDEIQQRVENGPCLAAAWEQHVIRIDDMESEQRWPAYCRQALLETPIRSVVAFQLYADNQTMGALNFYS ETAGAFDAEAVESGLIIATHAALMWSLMRRDEQFRSALASRDLIGQAKGMLMERYRIDAGQAFEVLKKLS QGANTPLVDVAREIVTTAERPADVSTQ*
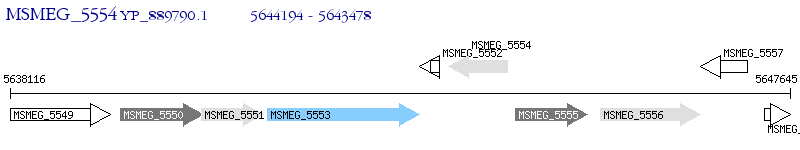
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5554 | - | - | 100% (238) | ANTAR domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_6741 | - | 1e-42 | 40.27% (226) | ANTAR domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_0672 | - | 1e-25 | 33.87% (186) | hypothetical protein MSMEG_0672 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3842 | - | 2e-61 | 50.45% (224) | response regulator receiver/ANTAR domain-containing protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2534c | - | 6e-47 | 42.01% (219) | hypothetical protein MAB_2534c |
| M. marinum M | MMAR_1808 | - | 1e-21 | 29.69% (192) | hypothetical protein MMAR_1808 |
| M. avium 104 | MAV_4102 | - | 6e-61 | 51.83% (218) | ANTAR domain-containing protein |
| M. thermoresistible (build 8) | TH_2526 | - | 4e-25 | 31.08% (251) | PUTATIVE putative response regulator receiver and ANTAR domain |
| M. ulcerans Agy99 | MUL_3601 | - | 1e-05 | 43.64% (55) | putative regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_2553 | - | 1e-63 | 50.44% (228) | response regulator receiver/ANTAR domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3842|M.gilvum_PYR-GCK MRAPRARGSGNLDVLLTRNTRTVTVNDSADVEAAHLRVAELVQGLHNRPD
Mvan_2553|M.vanbaalenii_PYR-1 ------------------------MSDPAGVEVTHLRVAELVQGLHNRPD
MSMEG_5554|M.smegmatis_MC2_155 -------------------------MSVSQNEAISQQISNLIRDVHTRRA
MAV_4102|M.avium_104 ------------------------MREAS--TTIVTQLVELVTSLE-REN
MAB_2534c|M.abscessus_ATCC_199 ------------------------MTDEAAAGAFSAKIADAARRMQ-QEH
TH_2526|M.thermoresistible__bu -------------------------------MGLNATVEKLIRKAR-ASG
MMAR_1808|M.marinum_M ---------------MTATPKDSNPDSVPGGTTADPATVFAALAEIIYQG
MUL_3601|M.ulcerans_Agy99 --------MSSPCHLPTPALSGQVPTVDGLQPSSQRSWCALPRALIGSAS
Mflv_3842|M.gilvum_PYR-GCK ADSDTVVAELAEHAAAEIPGAQYAGITLTRNAKHVETPAATHHWPLLLDK
Mvan_2553|M.vanbaalenii_PYR-1 TDSDTVVAELAELAAAEIEGAQYAGITLTRNAKQIETPAATHHWPLLLDK
MSMEG_5554|M.smegmatis_MC2_155 TDVDAVLGELTESAVEYVAGAQYAGITIAGRDGKVRTAAATGEYPEILDE
MAV_4102|M.avium_104 SDTAAGLHELIDNGVHHVTGSQYAGITLAEKSKSVNSVAATHRYPMVLDA
MAB_2534c|M.abscessus_ATCC_199 SSVEMTLRSITTAALETVPGVEQAGITLVTGRYEIESRAATSETPRKLDD
TH_2526|M.thermoresistible__bu ADTCAVLTELTRSAVGDWPSIQHAGITVVEDGDVVRSIAATDGHVLVVDN
MMAR_1808|M.marinum_M AQPAEMYSAICIAATLIVPGCDHASLLVRTN-DRYITVGASDRLAQHVDE
MUL_3601|M.ulcerans_Agy99 TLAQCGAGVQVRLGSWWLPGGSGAMISDVQQTAHTGEERPDRGTPSFATP
: . . . * : .
Mflv_3842|M.gilvum_PYR-GCK IQQRYLEGPCLTAA----------WEEQTVHVRDLQT-----EERFPNYR
Mvan_2553|M.vanbaalenii_PYR-1 IQQRYLEGPCLTAA----------WEEKTVHVRNLET-----DERFPNYR
MSMEG_5554|M.smegmatis_MC2_155 IQQRVENGPCLAAA----------WEQHVIRIDDMES-----EQRWPAYC
MAV_4102|M.avium_104 IQNKYGEGPCLAAA----------WEHHVMHIADLAA-----EQRWQRYR
MAB_2534c|M.abscessus_ATCC_199 IQQELREGPCVQSA----------WEHETVYAEDLAQ-----TTPWPRFA
TH_2526|M.thermoresistible__bu IARRLSTGPGIDAA----------LDEDSYRIDDLRA-----DSRWPLFA
MMAR_1808|M.marinum_M LERKAGDGPCIDAI----------EEETPQIETDLTT-----PTQWPKFA
MUL_3601|M.ulcerans_Agy99 ISGAVDNHLSIGSFRFWFIGQRWEWSDEVAGMHGYEPGAVVPTTKLLLSH
: : : .. .
Mflv_3842|M.gilvum_PYR-GCK RDALAETPIRSIMAFQL------------FIADESLGALNVYSEEPDVFG
Mvan_2553|M.vanbaalenii_PYR-1 RDALAETPIRSIMAFQL------------FIAGETLGALNVYSEQPEAFG
MSMEG_5554|M.smegmatis_MC2_155 RQALLETPIRSVVAFQL------------YADNQTMGALNFYSETAGAFD
MAV_4102|M.avium_104 RHALEQTPIRSILSFEL------------FVDRTSMAALNFYADPPHAFT
MAB_2534c|M.abscessus_ATCC_199 EAAVKLG-VRSMLSFQL------------FTFGENLGALNLYAQSPGAFG
TH_2526|M.thermoresistible__bu DKVAASTPIRSVIAIPVGNPIRNPIRNPIASAGTVRAALSLYSDLPGAFD
MMAR_1808|M.marinum_M AALLAQTPVRGAMGFRL------------LVDKRKGAALNLFSETPNSFD
MUL_3601|M.ulcerans_Agy99 KHPDDRAHVQDLLDYALRS------GESFSSRHRFVDTAGVVHDAIVVAD
::. : : : .. :
Mflv_3842|M.gilvum_PYR-GCK SEARTLGLIFAAHSSVAWNSARRE--------AQFRRALESRDVIGQAKG
Mvan_2553|M.vanbaalenii_PYR-1 DESRTLGLIFAAHSSVAWNSARRE--------EQFRQALSSRDVIGQAKG
MSMEG_5554|M.smegmatis_MC2_155 AEAVESGLIIATHAALMWSLMRRD--------EQFRSALASRDLIGQAKG
MAV_4102|M.avium_104 EESVELGTVYATHIALAWSMMRRQ--------DQFRSALASRDIIGQAKG
MAB_2534c|M.abscessus_ATCC_199 ADSYDAGLALATHAAIVLVAAQRE--------SQWASALASRDVIGQAKG
TH_2526|M.thermoresistible__bu DEVEAVAATYAEHIGRVLTARRGRRRAPVSGSARAQGSVRGQDAIETSKA
MMAR_1808|M.marinum_M TESAGRAAVLASFASVAVNAIAHG-----EDAASLRRGLLSNREIGKAVG
MUL_3601|M.ulcerans_Agy99 RMVNESGVVQGTSGYYIDLTNTFDETRQEVLDEALPDLFENRAAIEQAKG
. . . .. * : .
Mflv_3842|M.gilvum_PYR-GCK MIMERYRVDAVRAFELLRKLSQDSNVPLSKVAADLVAQAQTSNGQTG---
Mvan_2553|M.vanbaalenii_PYR-1 MIMERYRVDAVQAFELLRKLSQDSNVPLIKIATSLVSDAQASAD------
MSMEG_5554|M.smegmatis_MC2_155 MLMERYRIDAGQAFEVLKKLSQGANTPLVDVAREIVTTAERPADVSTQ--
MAV_4102|M.avium_104 VIMERFDLDAVEAFELLTRLSQQSNTRVVDIAAALIDSEHPLKQRRR---
MAB_2534c|M.abscessus_ATCC_199 MLMERFSINAAEAFTVIRTLSQDFNTKLVEIARAIVEDRPLG--------
TH_2526|M.thermoresistible__bu QLMKHFGVDPAAALTVLLRLSRECNRSLEDVARLLVGPRPVAEPPSQATA
MMAR_1808|M.marinum_M MLMLLNGFTEEEAFDLLRHHSQSLNIKLADVARTVIENRGQLPPEALETD
MUL_3601|M.ulcerans_Agy99 VLMAVYRVSAEEAFRVLQWRSQETNTKLRALAKQVVAEVASLPPVSALVQ
:* . *: :: *: * : :* ::
Mflv_3842|M.gilvum_PYR-GCK --------------------
Mvan_2553|M.vanbaalenii_PYR-1 --------------------
MSMEG_5554|M.smegmatis_MC2_155 --------------------
MAV_4102|M.avium_104 --------------------
MAB_2534c|M.abscessus_ATCC_199 --------------------
TH_2526|M.thermoresistible__bu TATPDHAGGRPPVWAEGAA-
MMAR_1808|M.marinum_M DTVNPTVSG-----------
MUL_3601|M.ulcerans_Agy99 SQFDHLLLTVHERIPAESAS