For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSIRRTVQEFVQRRTLLPDRIDSLIAGPGRDVRGLTVVVTGASAGIGRESVLQLRDQGARVIGVARRADE LKALAAETGCEWHTCDLSHQDQIARLLEELTEQGVDVLVNNAGHSIRRRIVDASDRLHDYQRTMALNYFA AVQLSLGLLPGMIERGRGQFVNVCTWALHANTFPRFSAYAASKSALAIFGRSLNAERPHPGVHATNVHFP LVRTEMITPTEEYNDAAALSAQEAGRWIMRAVTHQPVEISPAVLRAVLPTIDLLSPTASDSTIASLT
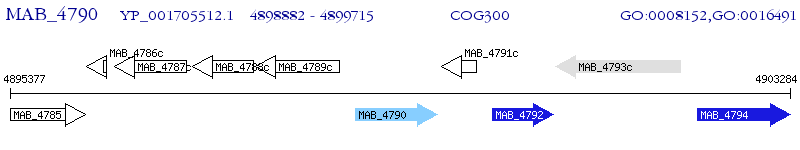
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4790 | - | - | 100% (277) | oxidoreductase |
| M. abscessus ATCC 19977 | MAB_3947 | - | 4e-52 | 46.98% (232) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3710 | - | 4e-37 | 38.11% (244) | short chain dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0561c | - | 1e-48 | 42.39% (243) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_0026 | - | 2e-50 | 40.07% (277) | short chain dehydrogenase |
| M. tuberculosis H37Rv | Rv0547c | - | 1e-48 | 42.39% (243) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_00429 | - | 7e-16 | 29.12% (261) | putative oxidoreductase |
| M. marinum M | MMAR_0894 | - | 6e-48 | 43.17% (227) | dehydrogenase |
| M. avium 104 | MAV_4597 | - | 3e-49 | 43.21% (243) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_1073 | - | 4e-50 | 44.26% (244) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_3707 | - | 2e-34 | 43.20% (169) | PUTATIVE oxidoreductase, short-chain dehydrogenase/reductase |
| M. ulcerans Agy99 | MUL_0647 | - | 4e-48 | 43.17% (227) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_0944 | - | 4e-52 | 40.43% (277) | short chain dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_1073|M.smegmatis_MC2_155 --------------------------MSKHPLRRLTDNLFLASMRPPVTD
TH_3707|M.thermoresistible__bu --------------------------------------------------
Mflv_0026|M.gilvum_PYR-GCK MRSSRKSASEVLLECEFGAGNRLRGVTSRNPLRRLADQVVLTSMRPPILP
Mvan_0944|M.vanbaalenii_PYR-1 -------------------------MTSRNPLRRLADQVVLTSMRPPILP
Mb0561c|M.bovis_AF2122/97 --------------------------MSKRPLRWLTEQITLAGMRPPISP
Rv0547c|M.tuberculosis_H37Rv --------------------------MSKRPLRWLTEQITLAGMRPPISP
MMAR_0894|M.marinum_M --------------------------MSKSPLRRLTEQITLAGMRPPIAP
MUL_0647|M.ulcerans_Agy99 --------------------------MSKSPLRRLTEQITLAGMRPPIAP
MAV_4597|M.avium_104 --------------------------MSKSPLRRFADQLVLATMRPPMAP
MAB_4790|M.abscessus_ATCC_1997 --------------------------MS---IRRTVQEFVQRRTLLPDRI
MLBr_00429|M.leprae_Br4923 --------------------------------------------------
MSMEG_1073|M.smegmatis_MC2_155 QLIQLRAARDAVDLTGKRVLITGASSGIGAAAARKFAERGARVIVVARRA
TH_3707|M.thermoresistible__bu -----------VRADGR------ADQGLDAARX-----------------
Mflv_0026|M.gilvum_PYR-GCK ALLN-RPTRETIELRGKRVLLTGASSGIGEAAAEKLARRGATVVAVARRQ
Mvan_0944|M.vanbaalenii_PYR-1 ALLN-RPNRETIELKGKRVLLTGASSGIGEAAAEKLARRGATVVAVARRG
Mb0561c|M.bovis_AF2122/97 QLLINRPAMQPVDLTGKRILLTGASSGIGAAATKQFGLHRAVVVAVARRK
Rv0547c|M.tuberculosis_H37Rv QLLINRPAMQPVDLTGKRILLTGASSGIGAAATKQFGLHRAVVVAVARRK
MMAR_0894|M.marinum_M QLLVNRPPIKPVALDGKRVMITGASSGIGEAAAEQFARQGATVIAVARRK
MUL_0647|M.ulcerans_Agy99 QLLVNRPPIKPVALDGKRVMITGASSGIGEAAAEQFARQGATVIAVARRK
MAV_4597|M.avium_104 QVLVNRPLIKPVELAGKRVLLTGASSGIGEAAAEQFAREGARVVVVARRK
MAB_4790|M.abscessus_ATCC_1997 DSLIAGPGR---DVRGLTVVVTGASAGIGRESVLQLRDQGARVIGVARRA
MLBr_00429|M.leprae_Br4923 -------MPIPAPSPEARAVVTGASQNIGEALATELAAHGHSLIVIARRE
*. .:.
MSMEG_1073|M.smegmatis_MC2_155 ELLDEVAGGITASG-GDAKARPCDLTDLDAIDALVAEIESDHGGLDILVN
TH_3707|M.thermoresistible__bu -------------G-GTGRALACDLTDLDAIDELVLKIEQDLGGVDILIN
Mflv_0026|M.gilvum_PYR-GCK DLLDELVTRIADAG-GDARAHACDLSDLDAVDELVATVEKDLGGIDILVN
Mvan_0944|M.vanbaalenii_PYR-1 DLLDDLVARITDAG-GRARAHACDLSDLDAIDALVATVEDELGGVDILVN
Mb0561c|M.bovis_AF2122/97 DLLDAVADRITGDG-GTAMSLPCDLSDMEAIDALVEDVEKRIGGIDILIN
Rv0547c|M.tuberculosis_H37Rv DLLDAVADRITGDG-GTAMSLPCDLSDMEAIDALVEDVEKRIGGIDILIN
MMAR_0894|M.marinum_M DLLDAVADRITTSG-GVAMSAPCDISDLDAVDALVADVENRLGGVDILIN
MUL_0647|M.ulcerans_Agy99 DLLDAVADRITTSG-GVAMSAPCDISDLDAVDALVADVENRLGGVDILIN
MAV_4597|M.avium_104 DLLDALAEGITRAG-GEAIAMPCDISDLDAADALVADVQQQLGGVDILIN
MAB_4790|M.abscessus_ATCC_1997 DELKALAAETG------CEWHTCDLSHQDQIARLLEELTEQ--GVDVLVN
MLBr_00429|M.leprae_Br4923 DVLVDLAARLADKYRVAVEVRPADLTDPSERVKLADELTVR--PISILCA
..*::. . * : :.:*
MSMEG_1073|M.smegmatis_MC2_155 NAGHSIRRPLADSLERWHDIERTMTLNYYSPLRLIRGFAPGMRERGAGHI
TH_3707|M.thermoresistible__bu NAGKSIRRPLAESLERWHDVERTMNLNYYAPLRLIRGLAPGMRE------
Mflv_0026|M.gilvum_PYR-GCK NAGRSIRRPLEESLERWHDVERTMTLNYYGPLRLIRGLAPGMLARGDGHI
Mvan_0944|M.vanbaalenii_PYR-1 NAGRSIRRPLEESLDRWHDVERTMTLNYYGPLRLIRGLAPGMLERGDGHV
Mb0561c|M.bovis_AF2122/97 NAGRSIRRPLAESLERWHDVERTMVLNYYAPLRLIRGLAPGMLERGDGHI
Rv0547c|M.tuberculosis_H37Rv NAGRSIRRPLAESLERWHDVERTMVLNYYAPLRLIRGLAPGMLERGDGHI
MMAR_0894|M.marinum_M NAGRSIRRPLAESLQRWHDVERTMVLNYYGPLRLIRGLAPGMLERGDGHI
MUL_0647|M.ulcerans_Agy99 NAGRSIRRPLAESLQRWHDVERTMVLNYYGPLRLIRGLAPGMLERGDGHI
MAV_4597|M.avium_104 NAGRSIRRPLAESLERWHDVERTMVLNYYAPLRLIRGIAPGMIERGDGHI
MAB_4790|M.abscessus_ATCC_1997 NAGHSIRRRIVDASDRLHDYQRTMALNYFAAVQLSLGLLPGMIERGRGQF
MLBr_00429|M.leprae_Br4923 NAGTATFGPVSALDP--AGEKAQVQLNAVAVHDLTLAVLPGMIERKAGGI
*** : : . : : ** . * .. ***
MSMEG_1073|M.smegmatis_MC2_155 INVSTWGVLSEASPLFSVYQASKAALSAVSRVIETEWGDGGVHSTTLYYP
TH_3707|M.thermoresistible__bu ----TWGVLSEASPLFSVYNASKAALTAVSRVVETEWSQFGVHSTTLFYP
Mflv_0026|M.gilvum_PYR-GCK INVSTWGVKTESPPLFGVYNASKAALSSVSRIIETEWSGRGVHATTLYYP
Mvan_0944|M.vanbaalenii_PYR-1 INVSTWGVKTESPPLFGVYNASKAALSSISRIMETEWADRGVHSTTLYYP
Mb0561c|M.bovis_AF2122/97 INVATWGVLSEASPLFSVYNASKAALSAVSRIIETEWGSQGVHSTTLYYP
Rv0547c|M.tuberculosis_H37Rv INVATWGVLSEASPLFSVYNASKAALSAVSRIIETEWGSQGVHSTTLYYP
MMAR_0894|M.marinum_M INVTTWGVLSEASPLFSVYNASKAALSAVSRTIETEWGRGGVHSTTLYYP
MUL_0647|M.ulcerans_Agy99 INVTTWGVLSEASPLFSVYNASKAALSAVSRTIETEWGRGGVHSTTLYYP
MAV_4597|M.avium_104 INVSTWGVLSEASPLFAVYNASKAALSTVSRVVETEWGDKGVHSTTLYYP
MAB_4790|M.abscessus_ATCC_1997 VNVCTWALHANTFPRFSAYAASKSALAIFGRSLNAERPHPGVHATNVHFP
MLBr_00429|M.leprae_Br4923 LISGSAAGNS-PIPYNATYAATKAFANTFSESLRGELHGSGVHVTLLAPG
: . : . * ..* *:*: ... :. * *** * :
MSMEG_1073|M.smegmatis_MC2_155 LVDTPMIKPTRA------FDGLPALSADEAADWMITAARHRPVQIAPRVA
TH_3707|M.thermoresistible__bu LVKTPMIAPTRE------YDNLPALSAEEAADWMISAARYRPIQIAPRMA
Mflv_0026|M.gilvum_PYR-GCK LVKTPMIAPTRA------YDGLPGLSADEAAGWIVTAARHRPVSIAPRIA
Mvan_0944|M.vanbaalenii_PYR-1 LVKTPMIAPTRA------YDGLPGLSAEEAAGWIITAARHRPVSIAPRIA
Mb0561c|M.bovis_AF2122/97 LVATPMIAPTKA------YDGLPALTAAEAAEWMVTAARTRPVRIAPRVA
Rv0547c|M.tuberculosis_H37Rv LVATPMIAPTKA------YDGLPALTAAEAAEWMVTAARTRPVRIAPRVA
MMAR_0894|M.marinum_M LVATPMIAPTKA------YSGMPALTPQEAAEWMITAARTRPVRIAPRIA
MUL_0647|M.ulcerans_Agy99 LVATPMIAPTKA------YSGMPALTPQEAAEWMITAARTRPVRIAPRIA
MAV_4597|M.avium_104 LVATPMIAPTKA------YQGVPALTPEEAGRWMITAARTRPVRIAPRMA
MAB_4790|M.abscessus_ATCC_1997 LVRTEMITPTEE------YNDAAALSAQEAGRWIMRAVTHQPVEISPAVL
MLBr_00429|M.leprae_Br4923 PVRTELPDHDEASLVEKLVPDFLWISTEHTARLSLDALERNKMRVVPGLT
* * : . . ::. .:. : * . : : * :
MSMEG_1073|M.smegmatis_MC2_155 VGAR-ALNAVAPGVVNAVMKRQSAQPDGVGRRPIFPAG
TH_3707|M.thermoresistible__bu VTAR-ALNVVAPGVVNAVMKRQRIQP-------VFV--
Mflv_0026|M.gilvum_PYR-GCK VTAH-AINSVAPGAVDTIMKRQRMQPRD----------
Mvan_0944|M.vanbaalenii_PYR-1 VTAH-AINSIAPAAVDTIMKRQRMQPKD----------
Mb0561c|M.bovis_AF2122/97 VAVN-ALDSIGPRWVNALMQRRNEQLNP----------
Rv0547c|M.tuberculosis_H37Rv VAVN-ALDSIGPRWVNALMQRRNEQLNP----------
MMAR_0894|M.marinum_M LMAK-ALNTLGPRWVNTLMQQRDDQLNDR---------
MUL_0647|M.ulcerans_Agy99 LMAK-ALNTLGPRWVNTLMQQRDDQLNDR---------
MAV_4597|M.avium_104 IAAK-ALDTFGPRWVNAVMQRQTVQPNREAGA------
MAB_4790|M.abscessus_ATCC_1997 RAVLPTIDLLSPTASDSTIASLT---------------
MLBr_00429|M.leprae_Br4923 SKAMSVASRYAPRAIVASIVGNFYKKLGGG--------
. . . .* : :