For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VKYIVTGGTGFIGRRIVTRILETQPAAEVAILVRRESLSRFEKLAEQWDDRVQPLVGDLTQPDLGLPAEG DPVTADHIVHCAAIYDITVDDKAQRAANVEGTRSVIALAKRTGAILHHISSIAVAGSYEGEFTEDDFDVN QDLPTPYHQTKFEAELLVRSEPGLRYRIYRPAVVVGDSRTGEMDKIDGPYYFFGVFAKLAKLPSFTPMML PRAGRSNIVPVDYVVDATVELMHQEGRDGQAFHLTNPEVTYLRDIYSGIAAAAGLPPVRGSLPHAVATPF LRARGTLKVWRNMVVTQLGIPPEVIDITEIMPTFISESTQEALRPSGIKVPPFGSYAGRLYQYWRRNLDP ERYRRDDPAGPLVGRHVIITGASSGIGRAAAIAVARRGGTVFAVARDGEALDQLVAEIREDGGKAHAFPC DLTDYAAVDDTVKSILGQFGHVDYLVNNAGRSIRRSVVNTVDRFHDYERVMAINYFGAVRLVLALLPHWR ERKFGHVVNVSTAGVQTRNPKYASYIPTKAALDAFADVVATETVSDHITFTNIHMPLVKTPMITPSHKLN VVRGLTPEHAAAMVVRGLIEKPTRIDHPMGTFADIGQYLTPKLSRRVLHQMYLAYPDSKAARGLEPVDDR RDLSTSRRRPRAAKRVAAVSRLRVPGPLMKAVRLIPGVHW
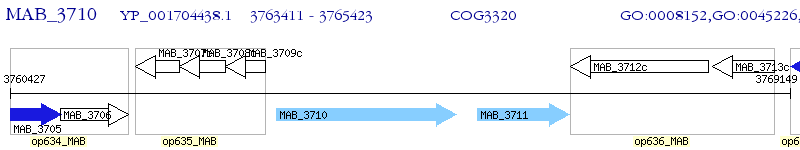
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3710 | - | - | 100% (670) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_2943c | - | 5e-47 | 40.30% (263) | putative oxidoreductase |
| M. abscessus ATCC 19977 | MAB_3947 | - | 1e-44 | 39.02% (264) | short chain dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3423 | acrA1 | 0.0 | 66.12% (670) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_4896 | - | 0.0 | 67.41% (675) | short chain dehydrogenase |
| M. tuberculosis H37Rv | Rv3391 | acrA1 | 0.0 | 66.12% (670) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_01094 | - | 4e-21 | 29.54% (237) | short chain alcohol dehydrogenase |
| M. marinum M | MMAR_1153 | acrA1 | 0.0 | 66.27% (673) | short-chain dehydrogenase, AcrA1 |
| M. avium 104 | MAV_4340 | - | 0.0 | 68.69% (677) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_1623 | - | 0.0 | 67.70% (678) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_3707 | - | 2e-27 | 36.11% (216) | PUTATIVE oxidoreductase, short-chain dehydrogenase/reductase |
| M. ulcerans Agy99 | MUL_0918 | acrA1 | 0.0 | 66.12% (673) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_1527 | - | 0.0 | 68.75% (656) | short chain dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4896|M.gilvum_PYR-GCK ----------MRYVVTGGTGFIGSRLIGRLLARPDVTAVHVLVRRGSLGR
Mvan_1527|M.vanbaalenii_PYR-1 --------------------------MTRLLSDPGTAAVHVLVRRDSLGR
MSMEG_1623|M.smegmatis_MC2_155 ----------MRYVVTGGTGFIGRRVVSEILVRDASAEVWVLVRRESLNR
Mb3423|M.bovis_AF2122/97 ----------MRYVVTGGTGFIGRHVVSRLLDGRPEARLWALVRRQSLSR
Rv3391|M.tuberculosis_H37Rv ----------MRYVVTGGTGFIGRHVVSRLLDGRPEARLWALVRRQSLSR
MAV_4340|M.avium_104 MRGRHRHDVFMRYVVTGGTGFIGRRVVTRLLQTRPDAQVWVLVRRQSLGR
MMAR_1153|M.marinum_M ----------MRYVVTGGTGFIGRRVVSRLLQSDADARVWVLVRRQSLSR
MUL_0918|M.ulcerans_Agy99 ----------MRYVVTGGTGFIGRRVVSRLLQSDADARVWVLVRRQSLSR
MAB_3710|M.abscessus_ATCC_1997 ----------MKYIVTGGTGFIGRRIVTRILETQPAAEVAILVRRESLSR
TH_3707|M.thermoresistible__bu --------------------------------------------------
MLBr_01094|M.leprae_Br4923 --------------------------------------------------
Mflv_4896|M.gilvum_PYR-GCK FERLARD----WDDRVHALVGDLTDPDLGLTDATVAELGPVDHVVHCAAI
Mvan_1527|M.vanbaalenii_PYR-1 FERLALN----WDDRVQPLVGELTVPGLGTSDVTVAELGRIDHVVHCAAV
MSMEG_1623|M.smegmatis_MC2_155 FEHLALD----WGERAKPLVGDLRADDLGLTGADVADLGDVTHVVHCAAI
Mb3423|M.bovis_AF2122/97 FERLAGQ----WGDRVRPLVGDL--TELELSERTIAELGDIDHVLHCAAV
Rv3391|M.tuberculosis_H37Rv FERLAGQ----WGDRVRPLVGDL--TELELSERTIAELGDIDHVLHCAAV
MAV_4340|M.avium_104 FERLSAQGDTPWGDRVKPLVGEL--PALELSDETLAELGHVDHLVHCAAI
MMAR_1153|M.marinum_M FERLASD----WGDRVKPLVGEL--PELPVTEQISAELGQIDHVVHCAAV
MUL_0918|M.ulcerans_Agy99 FERLATD----WGDRVKPLVGEL--PELPVTEQISAELGQIDHVVHCAAV
MAB_3710|M.abscessus_ATCC_1997 FEKLAEQ----WDDRVQPLVGDLTQPDLGLPAE--GDPVTADHIVHCAAI
TH_3707|M.thermoresistible__bu --------------------------------------------------
MLBr_01094|M.leprae_Br4923 --------------------------------------------------
Mflv_4896|M.gilvum_PYR-GCK YDMTAPENRQRAANVEGTRAVIALSRRLGATLHHVSSIAVAGTHPGVFTE
Mvan_1527|M.vanbaalenii_PYR-1 YDITAPEDRQRAANVDGTGAVVALSRRLGATLHHVSSIAVAGTYRGVFTE
MSMEG_1623|M.smegmatis_MC2_155 YDITVDEAAQRAANVDGTRAVIALARRLEATLHHVSSIAVAGNHRGVFTE
Mb3423|M.bovis_AF2122/97 HDTTW---------ADATRAVIELAARLDATFHHVSSIAVAGDFAGHYTE
Rv3391|M.tuberculosis_H37Rv HDTTW---------ADATRAVIELAARLDATFHHVSSIAVAGDFAGHYTE
MAV_4340|M.avium_104 YDITAGEAEQRAANVDGTRAVIELAQRLGATLHHVSSIAVAGDFPGEYTE
MMAR_1153|M.marinum_M YDITAGEDQQRAANVDGTRAVIELTRRLDATLHHVSSIAVAGDFAGEYTE
MUL_0918|M.ulcerans_Agy99 YDITAGEDQQRAANVDGTRAVIELTRRLDATLHHVSSIAVAGDFAGEYTE
MAB_3710|M.abscessus_ATCC_1997 YDITVDDKAQRAANVEGTRSVIALAKRTGAILHHISSIAVAGSYEGEFTE
TH_3707|M.thermoresistible__bu --------------------------------------------------
MLBr_01094|M.leprae_Br4923 --------------------------------------------------
Mflv_4896|M.gilvum_PYR-GCK DDVDVGQELPTPYHRTKFEAEQLVRAADGLRFRIYRPAVVVGDSRTGEID
Mvan_1527|M.vanbaalenii_PYR-1 DDFDVAQDLPTPYHRTKFEAELLVRSAEGLRFRIYRPAVVVGDSRTGEMD
MSMEG_1623|M.smegmatis_MC2_155 DDFDVAQDLPTPYHQTKFEAEMLVRATTDVPHRIYRPAVVVGDSRTGEMD
Mb3423|M.bovis_AF2122/97 ADFDVGQRLPTPYHRMTFEAERLVRSTPGLRYRIYRPAVVVGDSRTGEMD
Rv3391|M.tuberculosis_H37Rv ADFDVGQRLPTPYHRMTFEAERLVRSTPGLRYRIYRPAVVVGDSRTGEMD
MAV_4340|M.avium_104 DDFDVGQQLPTPYHQTKFEAELLVRSAPGLRHRIYRPAVVVGDSRTGEMD
MMAR_1153|M.marinum_M ADFDVGQQLPSPYHRTKFEAELLVRSSTGLRYRIYRPAVVVGDSRTGEMD
MUL_0918|M.ulcerans_Agy99 ADFDVGQQLPSPYHRTKFEAELLVRSSTGLRYRIYRPAVVVGDSRTGEMD
MAB_3710|M.abscessus_ATCC_1997 DDFDVNQDLPTPYHQTKFEAELLVRSEPGLRYRIYRPAVVVGDSRTGEMD
TH_3707|M.thermoresistible__bu --------------------------------------------------
MLBr_01094|M.leprae_Br4923 --------------------------------------------------
Mflv_4896|M.gilvum_PYR-GCK KIDGPYYFFGVLSALAALPRFTPMMLPDVGRTNLVPVDFVVDALDVLMHA
Mvan_1527|M.vanbaalenii_PYR-1 KIDGPYYFFGVLSALAALPGFTPMLLPDVGRTNIVPVDFVVEAVAALMHV
MSMEG_1623|M.smegmatis_MC2_155 KIDGPYYFFGVLARLAALPRFTPMALPDTGRTNVVPVDFVAKALVHLMHV
Mb3423|M.bovis_AF2122/97 TIDGPYYLFGVLAKLAVLPSFTPMLLPDIGRTNIVPVDYVADALVALMHA
Rv3391|M.tuberculosis_H37Rv TIDGPYYLFGVLAKLAVLPSFTPMLLPDIGRTNIVPVDYVADALVALMHA
MAV_4340|M.avium_104 KVDGPYYFFGVLAKLAVLPRFTPMLLPDTGRTNIVPVDYVADALVALMHA
MMAR_1153|M.marinum_M KIDGPYYFFGVLARLAALPSLTPILLPDTGRTNIVPVDYVVDALVALIHE
MUL_0918|M.ulcerans_Agy99 KIDGLYYFFGVLARLAALPSLTPILLPDTGRTNIVPVDYVVDALVALIHE
MAB_3710|M.abscessus_ATCC_1997 KIDGPYYFFGVFAKLAKLPSFTPMMLPRAGRSNIVPVDYVVDATVELMHQ
TH_3707|M.thermoresistible__bu --------------------------------------------------
MLBr_01094|M.leprae_Br4923 --------------------------------------------------
Mflv_4896|M.gilvum_PYR-GCK DGR---DGQTFHLTAPKNTGLRGIYRGVARAAGLPPLVGVLPRAAAAPLL
Mvan_1527|M.vanbaalenii_PYR-1 DGR---DGQTFHLTAPKTTGLRGIYRGIAAAAGLPPLVGSLPRATAAPLL
MSMEG_1623|M.smegmatis_MC2_155 EGA---DGQTFHLTAPNTVGLVDIYRGVAGAAGLPPLRAKLPRVTAAPVL
Mb3423|M.bovis_AF2122/97 DGR---DGQTFHLTAPTAIGLRGIYRGIAGAAGLPPLLGTLPGFVAAPVL
Rv3391|M.tuberculosis_H37Rv DGR---DGQTFHLTAPTAIGLRGIYRGIAGAAGLPPLLGTLPGFVAAPVL
MAV_4340|M.avium_104 DGL---DGQTFHLTAEKTIGLRGIYRGVAKAAGLPPLRGSLPGAVAAPVL
MMAR_1153|M.marinum_M DNMAFPNGRTFHLTAPQSVGLRGIYQAVAKEAGLPPARGALPGSVVTPVL
MUL_0918|M.ulcerans_Agy99 DNMAFPNGRTFHLTAPQSVGLRGIYQAVTKEAGLPPARGVLPGSVVTPVL
MAB_3710|M.abscessus_ATCC_1997 EGR---DGQAFHLTNPEVTYLRDIYSGIAAAAGLPPVRGSLPHAVATPFL
TH_3707|M.thermoresistible__bu --------------------------------------------------
MLBr_01094|M.leprae_Br4923 --------------------------------------------------
Mflv_4896|M.gilvum_PYR-GCK QVSGRAKAIRNMAATQLGIPGEVLDVVELAPTFTSERTDEALCDSGISVP
Mvan_1527|M.vanbaalenii_PYR-1 HVTGRAKVLRNMVATQLGIPAEILDIVELAPTFTSEHTDEALRGTGISVP
MSMEG_1623|M.smegmatis_MC2_155 DARGRARIVRNMVATQLGIPGEVFDVAELRPTFTADNTDTALRGTGIAVP
Mb3423|M.bovis_AF2122/97 NARGRAKVLRNMAATQLGIPAEIFDVVGCAPTFTSDTTREALRGTGIHVP
Rv3391|M.tuberculosis_H37Rv NARGRAKVLRNMAATQLGIPAEIFDVVGCAPTFTSDTTREALRGTGIHVP
MAV_4340|M.avium_104 KVRGRARVVRDMAATQLGIPAQVFDLVDLAPTFVSEKTRNALAGTGIEVP
MMAR_1153|M.marinum_M KARGRAKVLRNMAATQLGIPSEIFDVLDLKPTFISDETQKALRGSGVRLP
MUL_0918|M.ulcerans_Agy99 KARGRAKVLRNMAATQLGIPSEIFDVLDLKPTFISDETQKALRGSGVRLP
MAB_3710|M.abscessus_ATCC_1997 RARGTLKVWRNMVVTQLGIPPEVIDITEIMPTFISESTQEALRPSGIKVP
TH_3707|M.thermoresistible__bu --------------------------------------------------
MLBr_01094|M.leprae_Br4923 --------------------------------------------------
Mflv_4896|M.gilvum_PYR-GCK EFASYAPGLWRYWAEHLDPNRARQDDPAGPLVGRHVVITGASSGIGRASA
Mvan_1527|M.vanbaalenii_PYR-1 EFASYAPALWRYWAQHLDPDRARRDDPAGPLVGKHVIITGASSGIGRAAA
MSMEG_1623|M.smegmatis_MC2_155 EFASYAPQLFRYWAEHLDPDRARRCDPAGPLVGRHVIITGASSGIGRASA
Mb3423|M.bovis_AF2122/97 EFATYAPGLWRYWAEHLDPDRARRNDP---LLGRHVIITGASSGIGRASA
Rv3391|M.tuberculosis_H37Rv EFATYAPGLWRYWAEHLDPDRARRNDP---LLGRHVIITGASSGIGRASA
MAV_4340|M.avium_104 EFAGYAPRLWRYWAEHLDPDRARRDDPRGPLYGRHVIVTGASSGIGRASA
MMAR_1153|M.marinum_M DFAGYAPKLWRYWAQHLDPDRALRDDPRGPLVGRHVIITGASSGIGRASA
MUL_0918|M.ulcerans_Agy99 DFAGYAPKLWRYWAQHLDPDRALRDDPRGPLVGRHVIITGASSGIGRASA
MAB_3710|M.abscessus_ATCC_1997 PFGSYAGRLYQYWRRNLDPERYRRDDPAGPLVGRHVIITGASSGIGRAAA
TH_3707|M.thermoresistible__bu --------------------------------------------------
MLBr_01094|M.leprae_Br4923 ---------------------------MEGFAGKVAVVTGAGSGIGQALA
Mflv_4896|M.gilvum_PYR-GCK VAVAARGATVFALARNAEALDDLIAEIRESGGDAHAFTCDVTDPSAVEHT
Mvan_1527|M.vanbaalenii_PYR-1 IAVAARGATVFALARNAEALDDLIAEIRAAGGDAHAFTCDVTDSAAVEHT
MSMEG_1623|M.smegmatis_MC2_155 IAVARRGAVVFALARNGEALDELIAEIRADGGQAHAFTCDVTDSASVEHT
Mb3423|M.bovis_AF2122/97 IAVAKRGATVFALARNGNALDELVTEIRAHGGQAHAFTCDVTDSASVEHT
Rv3391|M.tuberculosis_H37Rv IAVAKRGATVFALARNGNALDELVTEIRAHGGQAHAFTCDVTDSASVEHT
MAV_4340|M.avium_104 IAIAQRGATVFALARNGAALDALVDEIRANGGAAHAFTCDVTDSASVEHT
MMAR_1153|M.marinum_M LAVAQRGGCVFALARNADALDQLVAEIRAGGGQAYAFTCDITDSTSVEHT
MUL_0918|M.ulcerans_Agy99 LAVAQRGGCVFALARNADALDQLVAEIRAGGGQAYAFTCDITDSTSVEHT
MAB_3710|M.abscessus_ATCC_1997 IAVARRGGTVFAVARDGEALDQLVAEIREDGGKAHAFPCDLTDYAAVDDT
TH_3707|M.thermoresistible__bu -------------VRADGRADQGLDAARX-GGTGRALACDLTDLDAIDEL
MLBr_01094|M.leprae_Br4923 IELARSGAKLAISDVDGEGLAQTEGQLKAIGASARTDRLDVTEREAFLTY
: *. . : *:*: :.
Mflv_4896|M.gilvum_PYR-GCK VKDILGRFGHVDYLVNNAGRSIRRSVTASTDRFHDYERVMAVNYFGAVRM
Mvan_1527|M.vanbaalenii_PYR-1 VKDILGRFDHVDYLVNNAGRSIRRSVAASTDRLHDYERVMAVNYFGSVRM
MSMEG_1623|M.smegmatis_MC2_155 VKDILGRFDHVDYLVNNAGRSIRRSVANSTDRLHDYERVMAVNYFGAVRM
Mb3423|M.bovis_AF2122/97 VKDILGRFDHVDYLVNNAGRSIRRSVVNSTDRLHDYERVMAVNYFGAVRM
Rv3391|M.tuberculosis_H37Rv VKDILGRFDHVDYLVNNAGRSIRRSVVNSTDRLHDYERVMAVNYFGAVRM
MAV_4340|M.avium_104 VKDILGRFDHVDYLVNNAGRSIRRSVVNSTDRLHDYERVMAVNYFGAVRM
MMAR_1153|M.marinum_M VKDILGRFGHVDYLVNNAGRSIRRSVVNSTDRLHDYERVMAVNYFGAVRM
MUL_0918|M.ulcerans_Agy99 VKDILGRFGHVDYLVNNAGRSIRRSVVNSTDRLHDYERVMAVNYFGAVRM
MAB_3710|M.abscessus_ATCC_1997 VKSILGQFGHVDYLVNNAGRSIRRSVVNTVDRFHDYERVMAINYFGAVRL
TH_3707|M.thermoresistible__bu VLKIEQDLGGVDILINNAGKSIRRPLAESLERWHDVERTMNLNYYAPLRL
MLBr_01094|M.leprae_Br4923 ADVVHENFGKVNQIYNNAGIAFTGDVEVS--HFKDIERVMDVDYWGVVNG
. : :. *: : **** :: : : : :* **.* ::*:. :.
Mflv_4896|M.gilvum_PYR-GCK VLALLPHWRERRFGHVVNVSSAGVQANNPRYSAYIPSKAALDAFSEVVGA
Mvan_1527|M.vanbaalenii_PYR-1 VLALLPHWRERRFGHVVNVSSAGVQANNPRYSAYIPSKAALDAFAEVVGT
MSMEG_1623|M.smegmatis_MC2_155 VLALLPHWRERRFGHVVNVSSAGVQAHTPKYSAYLPSKAALDAFSDVVST
Mb3423|M.bovis_AF2122/97 VLALLPHWRERRFGHVVNVSSAGVQARNPKYSSYLPTKAALDAFADVVAS
Rv3391|M.tuberculosis_H37Rv VLALLPHWRERRFGHVVNVSSAGVQARNPKYSSYLPTKAALDAFADVVAS
MAV_4340|M.avium_104 VLALLPHWRERRFGHVVNVSSAGVQARNPKYSSYLPTKAALDAFADVVGS
MMAR_1153|M.marinum_M VLALLPHWRERRFGHVVNVSSAGVQARNPKYSSYLPSKAALDAFADVVAS
MUL_0918|M.ulcerans_Agy99 VLALLPHWRERRFGHVVNVSSAGVQARNPKYSSYLPSKAALDAFADVVAS
MAB_3710|M.abscessus_ATCC_1997 VLALLPHWRERKFGHVVNVSTAGVQTRNPKYASYIPTKAALDAFADVVAT
TH_3707|M.thermoresistible__bu IRGLAPGMRETWG---------VLSEASPLFSVYNASKAALTAVSRVVET
MLBr_01094|M.leprae_Br4923 TKAFLPYLISSGDGHVINISSVFGLFSVPGQAAYNSAKFAVRGFTEALR-
.: * . * : * .:* *: ..: .:
Mflv_4896|M.gilvum_PYR-GCK ETLSDHITFTNIHMPLVKTPMIAPSGRLNPAPAISPEHAAAMVVRALVDK
Mvan_1527|M.vanbaalenii_PYR-1 ETLSDHITFTNIHMPLVRTPMIAPSGRLNPAPAISAEHAAAMVVRALVDK
MSMEG_1623|M.smegmatis_MC2_155 ETLSDHITFTNIHMPLVKTPMIAPSRRLNPVPPISAEHAAAMVVRALVEK
Mb3423|M.bovis_AF2122/97 ETLSDHITFTNIHMPLVATPMIVPSRRLNPVRAISAERAAAMVIRGLVEK
Rv3391|M.tuberculosis_H37Rv ETLSDHITFTNIHMPLVATPMIVPSRRLNPVRAISAERAAAMVIRGLVEK
MAV_4340|M.avium_104 EVLSDHITFTNIHMPLVRTPMIVPSHRLNPVRAISPERAAAMVVRGLVEK
MMAR_1153|M.marinum_M ETLSDHVTFTNIHMPLVATPMIVPSRRLNPTPAISAEHAAAMVVRGLVEK
MUL_0918|M.ulcerans_Agy99 ETLSDHVTFTNIHMPLVATPMIVPSRRLNPTPAISAEHAAAMVVRGLVEK
MAB_3710|M.abscessus_ATCC_1997 ETVSDHITFTNIHMPLVKTPMITPSHKLNVVRGLTPEHAAAMVVRGLIEK
TH_3707|M.thermoresistible__bu EWSQFGVHSTTLFYPLVKTPMIAPTREYDNLPALSAEEAADWMISAARYR
MLBr_01094|M.leprae_Br4923 -----------EEMALAGRPVNVTTVYPGGIKTAIARNATAAEGLDVSKI
.*. *: ..: . ...*:
Mflv_4896|M.gilvum_PYR-GCK PARIDTPIGTVADLGMYLTPKLARRVLHQLYLGFPDSAAAQGIAENDRVH
Mvan_1527|M.vanbaalenii_PYR-1 PARIDTPIGTLADVGMYFTPKLARRVLHQLYLGFPDSAAARGITQ-DRSR
MSMEG_1623|M.smegmatis_MC2_155 PPRIDTPLGTFADFGNYLTPRVARRFLHQLYLGYPDSAAARGLAPAEPRP
Mb3423|M.bovis_AF2122/97 PARIDTPLGTLAEAGNYVAPRLSRRILHQLYLGYPDSAAAQGISRPD--A
Rv3391|M.tuberculosis_H37Rv PARIDTPLGTLAEAGNYVAPRLSRRILHQLYLGYPDSAAAQGISRPD--A
MAV_4340|M.avium_104 PARIDTPLGTLAEAGNYFAPRTSRRVLHQLYLGYPDSAAARGVATEPRPA
MMAR_1153|M.marinum_M PARIDTPLGTLAEVGNYVAPKLSRRILHQLYLGYPDSAAAQGISLLD---
MUL_0918|M.ulcerans_Agy99 PARIDTPLGTLAEIGNYVAPKLSRRILHQLYLGYPDSAAAQGISPLN---
MAB_3710|M.abscessus_ATCC_1997 PTRIDHPMGTFADIGQYLTPKLSRRVLHQMYLAYPDSKAARGLEPVDDRR
TH_3707|M.thermoresistible__bu PIQIAPRMAVTARALNVVAPGVVNAVMKRQRIQPVFV-------------
MLBr_01094|M.leprae_Br4923 ASRFDTWVAHTSPQHAARIILKAVRKKKARVLVGPDAKVANVVVRFSGGA
. :: :. : : :
Mflv_4896|M.gilvum_PYR-GCK SATRTP---ERKPWPPRRPVPSVRIP----SAVKQAIRLVPGVHW
Mvan_1527|M.vanbaalenii_PYR-1 -PRQAP---VRRPKRPSRPMPSVRVP----GPVKRAARLLPGLHW
MSMEG_1623|M.smegmatis_MC2_155 ASTAGPGLPERRPKRPARSRRGVRVPRAVGRPVKRAVRTLPGVHW
Mb3423|M.bovis_AF2122/97 DRPP-------APRRPRRSARAG-----VPRPLRRLGRLVPGVHW
Rv3391|M.tuberculosis_H37Rv DRPP-------APRRPRRSARAG-----VPRPLRRLGRLVPGVHW
MAV_4340|M.avium_104 ERIERT-----APRKPRRPVRAVTRGLRTPRPVRRLVRLVPGVHW
MMAR_1153|M.marinum_M DAQEQR-----AVRRPKRPALAVTG-MRIPRPVKRLVRLTPGVHW
MUL_0918|M.ulcerans_Agy99 DAQEHR-----PVRRPKRPALAVTG-MRIPRPVKRLVRLAPGVHW
MAB_3710|M.abscessus_ATCC_1997 DLSTSR----RRPRAAKRVAAVSRLR--VPGPLMKAVRLIPGVHW
TH_3707|M.thermoresistible__bu ---------------------------------------------
MLBr_01094|M.leprae_Br4923 GYQRLF--------------------------AQVASRLILNQR-