For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RFAFKTSPQNTTWDDMLAIWKVADDIEVFESGWTFDHFYPIFSDPTGPCLEGWVTLTALAQATQRLRVGV LVTGIHYRHPAVLANMASALDIVSGGRLELGIGAGWNEEESGAYGIELGSIKERFDRFEEACEVLTGLLS RETTNFDGKFYQLKDARNEPKGPQKPHPPICIGGSGEKRTLRITARYAQHWNFVGGPPEEFARKREVLAA HCADIGRDPKEITLSAHIRLGADRDYARVVDEAASLGAEGLDLAIIYLPPPYDPAVLEPLAQALRG*
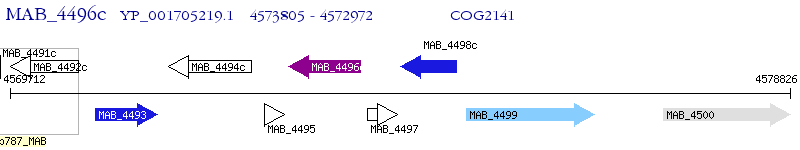
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4496c | - | - | 100% (277) | luciferase-like monooxygenase |
| M. abscessus ATCC 19977 | MAB_4797 | - | e-104 | 63.51% (285) | luciferase-like monooxygenase |
| M. abscessus ATCC 19977 | MAB_2431c | - | 3e-44 | 42.54% (228) | luciferase-like oxidoreductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1886c | - | 4e-41 | 38.66% (269) | oxidoreductase |
| M. gilvum PYR-GCK | Mflv_1117 | - | 1e-146 | 87.32% (276) | luciferase family protein |
| M. tuberculosis H37Rv | Rv1855c | - | 4e-41 | 38.66% (269) | oxidoreductase |
| M. leprae Br4923 | MLBr_00131 | - | 4e-14 | 33.50% (200) | putative oxidoreductase |
| M. marinum M | MMAR_5411 | - | 1e-140 | 81.88% (276) | hypothetical protein MMAR_5411 |
| M. avium 104 | MAV_0169 | - | 1e-143 | 84.06% (276) | hypothetical protein MAV_0169 |
| M. smegmatis MC2 155 | MSMEG_6454 | - | 1e-150 | 89.49% (276) | hypothetical protein MSMEG_6454 |
| M. thermoresistible (build 8) | TH_4285 | - | 1e-142 | 83.70% (276) | PUTATIVE luciferase family protein |
| M. ulcerans Agy99 | MUL_5034 | - | 1e-139 | 81.88% (276) | hypothetical protein MUL_5034 |
| M. vanbaalenii PYR-1 | Mvan_5695 | - | 1e-146 | 87.32% (276) | luciferase family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1886c|M.bovis_AF2122/97 ---MTIRLGLQIPNFSYGTGVEKLFPSVIAQAREAEAAGYDSLFVMDHFY
Rv1855c|M.tuberculosis_H37Rv ---MTIRLGLQIPNFSYGTGVEKLFPSVIAQAREAEAAGYDSLFVMDHFY
MAB_4496c|M.abscessus_ATCC_199 ---MRFAFKTSPQNTTWDD-------MLAIWKVADDIEVFESGWTFDHFY
MSMEG_6454|M.smegmatis_MC2_155 ---MRFAFKTSPQNTTWAD-------MLAIWKAADNIDVFESGWTFDHFY
Mflv_1117|M.gilvum_PYR-GCK ---MRFAFKTSPQNTTWSD-------MLTVWKAADDIDVFESGWTFDHFY
Mvan_5695|M.vanbaalenii_PYR-1 ---MRFAFKTSPQNTAWSD-------MLSVWKAADDIDVFESGWTFDHFY
TH_4285|M.thermoresistible__bu ---MQFGFKTSPQNTTWQD-------MLSVWRAADDIDVFESGWTFDHFY
MMAR_5411|M.marinum_M ---MRFAFKTSPQNTTWEQ-------MLAVWQTADGIDVYESGWTFDHFY
MUL_5034|M.ulcerans_Agy99 ---MRFAFKNSPQNTTWEQ-------MLAVWQTADGIDVYESGWTFDHFY
MAV_0169|M.avium_104 ---MRFAFKTSPQNTTWAQ-------MLAVWQAADDIDVYESGWTFDHFY
MLBr_00131|M.leprae_Br4923 MAGFRFGFVDALVHTLFPP-----SLPARASIVSGAVLGADSYWVGDHLN
: : : : : :* :. **:
Mb1886c|M.bovis_AF2122/97 QLPMLGTPD--------------QPMLEAYTALGALATATER--LQLGAL
Rv1855c|M.tuberculosis_H37Rv QLPMLGTPD--------------QPMLEAYTALGALATATER--LQLGAL
MAB_4496c|M.abscessus_ATCC_199 --PIFSDPT--------------GPCLEGWVTLTALAQATQR--LRVGVL
MSMEG_6454|M.smegmatis_MC2_155 --PIFSDSA--------------GPCLEGWTTLTALAQATER--LRVGVL
Mflv_1117|M.gilvum_PYR-GCK --PIFSDPT--------------GPCLEGWVTLTALAQATTR--LRVGVL
Mvan_5695|M.vanbaalenii_PYR-1 --PIFSDPS--------------GPCLEGWVTLTALAQATTR--LRVGVL
TH_4285|M.thermoresistible__bu --PIFSDPT--------------GPCLEGWTTLTALAQATTR--LRVGVL
MMAR_5411|M.marinum_M --PIFSDPT--------------GPCLEGWTTLTALAQATTR--LRLGTL
MUL_5034|M.ulcerans_Agy99 --PIFSDPT--------------GPCLEGWTTLTALAQATTR--LRLGTL
MAV_0169|M.avium_104 --PIFSDSS--------------GPCLEGWTTLTALAQATKR--LRLGTL
MLBr_00131|M.leprae_Br4923 ALVPRSVATPKYLGVAAKVVPKIDANYEPWTMLGNLAAGNRLNGLRLGVC
. . . * :. * ** .. *::*.
Mb1886c|M.bovis_AF2122/97 VTGNTYRSPTLLAKIITTLDVVSAGRAILGIGAGWFELEHRQLGFEFGTF
Rv1855c|M.tuberculosis_H37Rv VTGNTYRSPTLLAKIITTLDVVSAGRAILGIGAGWFELEHRQLGFEFGTF
MAB_4496c|M.abscessus_ATCC_199 VTGIHYRHPAVLANMASALDIVSGGRLELGIGAGWNEEESGAYGIELGSI
MSMEG_6454|M.smegmatis_MC2_155 VTGIHYRHPAVLANMAAALDIISNGRLELGIGAGWNEEESGAYGIELGSI
Mflv_1117|M.gilvum_PYR-GCK VTGIHYRHPAVLANMASALDIVSGGRLELGIGAGWNEEESGAYGIELGSI
Mvan_5695|M.vanbaalenii_PYR-1 VTGIHYRHPAVLANMASALDIVSDGRLELGIGAGWNEEESGAYGIELGSI
TH_4285|M.thermoresistible__bu VTGIHYRHPAVLANMASALDIISAGRLELGIGAGWNEEESGAYGIELGTL
MMAR_5411|M.marinum_M VTGVHYRHPAVLANMAAALDIISHGRLELGIGAGWNEEESSAYGIELGSV
MUL_5034|M.ulcerans_Agy99 VTGIHYRHPAVLANMAAALDIISHGRLELGIGAGWNEEESGAYGIELGSV
MAV_0169|M.avium_104 VTGIHYRHPAVLANMAAAVDIISGGRLELGIGAGWNEEESGAYGIELGSI
MLBr_00131|M.leprae_Br4923 VTDAGRRNPAVTAQAAATLHLLTRGKAMLGIGVGERE-GNEPYGVEW---
**. * *:: *: :::.::: *: ****.* * *.*
Mb1886c|M.bovis_AF2122/97 SDRFNRLEEALQILEPMVK--GERPTFFGDWYTTESAMAEPR--YRDRIP
Rv1855c|M.tuberculosis_H37Rv SDRFNRLEEALQILEPMVK--GERPTFFGDWYTTESAMAEPR--YRDRIP
MAB_4496c|M.abscessus_ATCC_199 KERFDRFEEACEVLTGLLS--RETTNFDGKFYQLKDARNEPKGPQKPHPP
MSMEG_6454|M.smegmatis_MC2_155 KERFDRFEEACEVLKGLLT--QETTTFDGKFYQLKDARNEPKGPQQPHPP
Mflv_1117|M.gilvum_PYR-GCK TERFDRFEEACQVLRGLLS--QETTTSDGRFYQLKDARNEPKGPQQPHPP
Mvan_5695|M.vanbaalenii_PYR-1 KERFDRFEEACQVLKGLLS--QDTTTFDGKFYQLKDARNEPKGPQRPHPP
TH_4285|M.thermoresistible__bu KERFDRFEEACQVLISLLS--QETTDFDGAYYRLKSARNEPKGPQQPHPP
MMAR_5411|M.marinum_M KERFDRFEEACAVLTGLLS--QPSTTFDGKYYQLEDARNEPKGPQRPHPP
MUL_5034|M.ulcerans_Agy99 KERFDRFEEACAVLTGLLS--QPSTTFDGKYYQLEDARNEPKGPQRPHSP
MAV_0169|M.avium_104 RERFDRFEEACAVLTSLLS--QETTDFDGKFYQLKGARNEPKGPQRPHPP
MLBr_00131|M.leprae_Br4923 TKPVARFQEALATIRALWDSNGELVSRESQFFPLHNALFDLPPYRGKWPE
. . *::** : : . :: ..* :
Mb1886c|M.bovis_AF2122/97 ILIGGGGEKKTFAIAARFADHLN-IVA-AVDELPRKMRALAARCDEAGRD
Rv1855c|M.tuberculosis_H37Rv ILIGGGGEKKTFAIAARFADHLN-IVA-AVDELPRKMRALAARCDEAGRD
MAB_4496c|M.abscessus_ATCC_199 ICIGGSGEKRTLRITARYAQHWN-FVGGPPEEFARKREVLAAHCADIGRD
MSMEG_6454|M.smegmatis_MC2_155 FCIGGSGEKRTLRITAKYADHWN-FVGGTPEEFARKRDVLAAHCADLGRD
Mflv_1117|M.gilvum_PYR-GCK ICIGGNGEKRTLRITAQYADHWN-YVGGTPEEFARKRDVLAAHCADIGRD
Mvan_5695|M.vanbaalenii_PYR-1 ICIGGSGEKRTLRITAQYADHWN-FVGGPPEEFARKRDVLARHCADIGRD
TH_4285|M.thermoresistible__bu ICIGGSGEKRTLRITAKYADHWN-FVGGPPEEFARKRDVLAAHCADVGRD
MMAR_5411|M.marinum_M ICIGGSGEKRTLRITARYAQHWN-FAGGTPEEFARKRDVLAAHCADIGRD
MUL_5034|M.ulcerans_Agy99 ICIGGSGEKRTLRITARYAQHWN-FAGGTPEEFARKRDVLAAHCADIGRD
MAV_0169|M.avium_104 ICIGGSGEKRTLRITARYAQHWN-FVGGPPDVFAHKRDVLASHCADIGRD
MLBr_00131|M.leprae_Br4923 IWVAAHG-PRMLQATGRYADAWIPIVLVRPTDYSCALEVVRTAASDAGRD
: :.. * : : :.::*: . . .: . : ***
Mb1886c|M.bovis_AF2122/97 RSTLQTSLLLTVMIDETLSPDAIPAEMSGRVVVGSPAQIADQIQA-KVLD
Rv1855c|M.tuberculosis_H37Rv RSTLQTSLLLTVMIDETLSPDAIPAEMSGRVVVGSPAQIADQIQA-KVLD
MAB_4496c|M.abscessus_ATCC_199 PKEITLSAHIRLGADRDYA-----------RVVDEAASLG---------A
MSMEG_6454|M.smegmatis_MC2_155 PKEITLSAHVRLGEDRDYA-----------KVIDEAAALG---------A
Mflv_1117|M.gilvum_PYR-GCK PKEIMLSAHVRLGADLDVD-----------KVIAEAAALG---------D
Mvan_5695|M.vanbaalenii_PYR-1 PGEISLSAHIRLGADLDYD-----------KVIEEAAALG---------A
TH_4285|M.thermoresistible__bu PREIRLSAHVRLGEDGDCR-----------KVVDEAAALA---------S
MMAR_5411|M.marinum_M PKEITLSAHLGLAPERDFG-----------QVIENAAALA---------A
MUL_5034|M.ulcerans_Agy99 PKEITLSAHRGLAPERDFG-----------QVIENAAALA---------A
MAV_0169|M.avium_104 PKEILLSAHLRLEPDLDYG-----------KVIDDAAALA---------A
MLBr_00131|M.leprae_Br4923 PMSITPAAVRGIITGRTRDDVDEALDSVLVRMIALGVPGEAWARHGVEHP
: : : :: .
Mb1886c|M.bovis_AF2122/97 AGVDGL-IINLAPHGYLPGVITTAAEALRPLLGV----------------
Rv1855c|M.tuberculosis_H37Rv AGVDGL-IINLAPHGYLPGVITTAAEALRPLLGV----------------
MAB_4496c|M.abscessus_ATCC_199 EGLDLA-IIYLPPP-YDPAVLEPLAQALRG--------------------
MSMEG_6454|M.smegmatis_MC2_155 EGLDLA-IIYLPPP-YDPAVLEPLAQAIRDSGQLTAA-------------
Mflv_1117|M.gilvum_PYR-GCK EGLDLA-IVYLPPP-YDPAVLEPLAEKIRASGLLDSNVDG----------
Mvan_5695|M.vanbaalenii_PYR-1 EGLELA-IVYLPLP-HDPAVLEPLADKIRASGLLDSKAR-----------
TH_4285|M.thermoresistible__bu EGLDLA-IVYLPTP-HTPAVLEPLAEAIRDSGLPAR--------------
MMAR_5411|M.marinum_M EGLDLG-IVYIAPP-HDPAVLEPMAEAIRESGLIDG--------------
MUL_5034|M.ulcerans_Agy99 EGLDLG-IVYIAPP-HDPAVLEPMAEAIRESGLIDG--------------
MAV_0169|M.avium_104 EGLDLG-IVYLPPP-HDPAVLEPLAEAIRDSGLLRS--------------
MLBr_00131|M.leprae_Br4923 MGADFAGVQDIIPQTIDEETVVSYAAKVPAALMKEVLFSGTPEEVIDQVA
* : : : .: . * :
Mb1886c|M.bovis_AF2122/97 ------------------------------------------
Rv1855c|M.tuberculosis_H37Rv ------------------------------------------
MAB_4496c|M.abscessus_ATCC_199 ------------------------------------------
MSMEG_6454|M.smegmatis_MC2_155 ------------------------------------------
Mflv_1117|M.gilvum_PYR-GCK ------------------------------------------
Mvan_5695|M.vanbaalenii_PYR-1 ------------------------------------------
TH_4285|M.thermoresistible__bu ------------------------------------------
MMAR_5411|M.marinum_M ------------------------------------------
MUL_5034|M.ulcerans_Agy99 ------------------------------------------
MAV_0169|M.avium_104 ------------------------------------------
MLBr_00131|M.leprae_Br4923 EWRDHGLKYLVVINGSLVNSSLRKTVSALLPHARVLRGLKKL