For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSISTYGVDYPASIDFIQAGKGSDDLSKHIQKTAADCPKMKFVVGGYAQGAAVVDVLLGNTPPGTYTFTN PLPAGLEDRIVAIVLFGDPKNIRPPGVDANLAIRDELLHKVIDVCNPGDFFCDPQGGDIISHTTYAKDKL TDGAADTVVQRLVALLGHRCKGADGQSTDSATTCSPTA
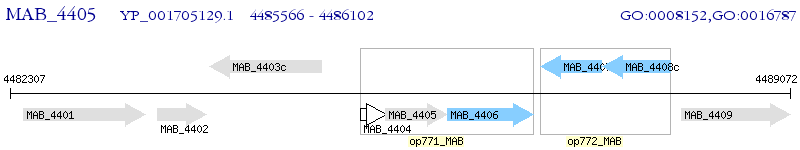
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4405 | - | - | 100% (178) | serine esterase cutinase family |
| M. abscessus ATCC 19977 | MAB_3809c | - | 9e-44 | 46.89% (177) | cutinase Cut4 |
| M. abscessus ATCC 19977 | MAB_3272c | - | 3e-22 | 35.58% (163) | cutinase Cut1 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3482 | cut4 | 5e-24 | 41.72% (151) | cutinase precursor CUT4 |
| M. gilvum PYR-GCK | Mflv_4966 | - | 2e-23 | 39.10% (156) | cutinase |
| M. tuberculosis H37Rv | Rv3452 | cut4 | 5e-24 | 41.72% (151) | cutinase precursor CUT4 |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_1097 | cut4 | 1e-23 | 39.74% (151) | cutinase precursor, Cut4 |
| M. avium 104 | MAV_4396 | - | 4e-26 | 42.48% (153) | cutinase |
| M. smegmatis MC2 155 | MSMEG_6354 | - | 2e-23 | 35.37% (164) | serine esterase, cutinase family protein |
| M. thermoresistible (build 8) | TH_4127 | - | 3e-19 | 37.58% (149) | PUTATIVE serine esterase, cutinase family protein |
| M. ulcerans Agy99 | MUL_0855 | cut4 | 3e-23 | 39.07% (151) | cutinase precursor, Cut4 |
| M. vanbaalenii PYR-1 | Mvan_1441 | - | 1e-23 | 40.40% (151) | cutinase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3482|M.bovis_AF2122/97 -----MIPRPQPHSGRWRAGAARRLTSLVAAAFAAATLLLTPALAPPAS-
Rv3452|M.tuberculosis_H37Rv -----MIPRPQPHSGRWRAGAARRLTSLVAAAFAAATLLLTPALAPPAS-
MMAR_1097|M.marinum_M MWLFPLDAQKWARSGRGRTRLARRCAGLVATTIAACAVLLAFPLVPKAA-
MUL_0855|M.ulcerans_Agy99 MWLFPLDAQKWARSGRGRTRLARRCAGLVATTIAACAVLLAFPLVPKAA-
MAV_4396|M.avium_104 -------------------------MGLGVGLLAGAAVLIAPQLIPLAS-
Mvan_1441|M.vanbaalenii_PYR-1 ----MMIDLDAP---------ARRATGVLAAILVAAATLVAAPVQIPAAH
Mflv_4966|M.gilvum_PYR-GCK --------------------------------MVPMAALAVMPTTTASA-
TH_4127|M.thermoresistible__bu -------------------------------VLGSAALLAGVPAPTAAA-
MSMEG_6354|M.smegmatis_MC2_155 -----MFKSTLSR------FIPAVSAALLTAGLVPGTAGTANAEPPIEGA
MAB_4405|M.abscessus_ATCC_1997 --------------------------------------------------
Mb3482|M.bovis_AF2122/97 -AGCPDAEVVFARGTGEPPGLGRVGQAFVSSLRQQTN-KSIGTYGVNYPA
Rv3452|M.tuberculosis_H37Rv -AGCPDAEVVFARGTGEPPGLGRVGQAFVSSLRQQTN-KSIGTYGVNYPA
MMAR_1097|M.marinum_M -AACPDVQVVFARGTGEPPGLGRVGNAMVASLRQKTD-QSIGAYAVNYPA
MUL_0855|M.ulcerans_Agy99 -AACPDVQVVFARGTGEPPGLGRVGNAMVASLRQKTD-QSIGAYAVNYPA
MAV_4396|M.avium_104 -ASCPGVEVVFARGTDEPPGVGKVGGAFVSSLREQTR-KSVGAYGVNYPA
Mvan_1441|M.vanbaalenii_PYR-1 AQGCPDIEVVFARGTDEPPGLGRVGAAFVDSLRGRVGGRSVGTYAVNYPA
Mflv_4966|M.gilvum_PYR-GCK -QPCPDVEVVFARGTSEPPGVGRVGQALADQLRNQLGGRTVGTYGVVYPA
TH_4127|M.thermoresistible__bu -IPCSDVEVVFARGTSEPPGVGRVGQALANALQARLPGRTVSTYGVSYPA
MSMEG_6354|M.smegmatis_MC2_155 APPCPDVEVVFARGTFEPPGVGFVGQAFVDALRGRLGDKSVDVYAVNYPA
MAB_4405|M.abscessus_ATCC_1997 --------------------------------------MSISTYGVDYPA
::..*.* ***
Mb3482|M.bovis_AF2122/97 NGDFLAAADGANDASDHIQQMASACRATRLVLGGYSQGAAVIDIVTAAPL
Rv3452|M.tuberculosis_H37Rv NGDFLAAADGANDASDHIQQMASACRATRLVLGGYSQGAAVIDIVTAAPL
MMAR_1097|M.marinum_M NKDFLAAADGANDASDHVQQMTNECPNTKLVLGGYSQGAAVIDIVTAAPL
MUL_0855|M.ulcerans_Agy99 NKDFLAAADGANDASDHVQQMTNECPNTKLVLGGYSQGAAVINIVTAAPL
MAV_4396|M.avium_104 NKDFLAAADGANDASNHIQQMAGNCPNTKLVLGGYSQGAAVIDIVTAAPV
Mvan_1441|M.vanbaalenii_PYR-1 SFDFLAAAAGANDASLHIQYMMANCPRTRLVLGGYSQGAAVIDVIAAVPI
Mflv_4966|M.gilvum_PYR-GCK SYDFFTSGDGAADATNRISVMAAQCPNTRIVLGGYSQGAAVVDMLAGVPP
TH_4127|M.thermoresistible__bu TYDFLAATDGANDATARITTLAQQCPSTRVVLGGYSQGAAVVDMLGGLPP
MSMEG_6354|M.smegmatis_MC2_155 SLDFQRAADGIVDASRKVEQTAATCPSTKTVIGGFSQGAAVAAYLTADSV
MAB_4405|M.abscessus_ATCC_1997 SIDFIQAGKGSDDLSKHIQKTAADCPKMKFVVGGYAQGAAVVDVLLGNTP
. ** : * * : :: * : *:**::***** : . .
Mb3482|M.bovis_AF2122/97 PG-----LGFTQPLPPAADDHIAAIALFGNPSGRAGGLMSALTP------
Rv3452|M.tuberculosis_H37Rv PG-----LGFTQPLPPAADDHIAAIALFGNPSGRAGGLMSALTP------
MMAR_1097|M.marinum_M PG-----LGFTQPLPAQAAQHVAAVTLFGNPSGRAGGLMTALSP------
MUL_0855|M.ulcerans_Agy99 PG-----LGFTQPLPAQAAQHVAAVTLFGNPSGRAGGLMTALSP------
MAV_4396|M.avium_104 PG-----LGFRNPLPPQAADHVAAVALFGNPSGRAGRLMSALTP------
Mvan_1441|M.vanbaalenii_PYR-1 PA-----VGFNAPLPPNTPDFVAAIAVFGNPSTKLG-IPITASP------
Mflv_4966|M.gilvum_PYR-GCK LGNRIGDVGLARPLPGGLNGNVAAVAVFGNPATKFG-NPVSARG------
TH_4127|M.thermoresistible__bu LGDRVGLVGSA--LPTGSVGNVAAVAVFGNPSAKFG-RPINTSP------
MSMEG_6354|M.smegmatis_MC2_155 PANFVLPDGITGPMPDDVAKHVTAVTLFGKPSNGFINTVQSTAPPINIGH
MAB_4405|M.abscessus_ATCC_1997 PG----TYTFTNPLPAGLEDRIVAIVLFGDPKNIRPPGVDANLAIR---D
. :* :.*:.:**.*
Mb3482|M.bovis_AF2122/97 QFGSKTINLCNNGDPICS-DGNRWRAHLGYVPG-MTNQAARFVASRI---
Rv3452|M.tuberculosis_H37Rv QFGSKTINLCNNGDPICS-DGNRWRAHLGYVPG-MTNQAARFVASRI---
MMAR_1097|M.marinum_M HFGGKILNLCNTGDPICS-DGDQWKAHTGYVPG-LTNKAASFVAGRI---
MUL_0855|M.ulcerans_Agy99 HFGGKILNLCNTGDPICS-DGDQWKAHTGYVPG-LTNKAASFVAGRI---
MAV_4396|M.avium_104 DFSGKTIDLCNPGDPICS-GGYQWASHLSYVPG-LTNQAARFVAARV---
Mvan_1441|M.vanbaalenii_PYR-1 VWGSRSIDLCNGLDPVCS-GGDDIAAHSNYGPGMFTDQAAAFVAGRL---
Mflv_4966|M.gilvum_PYR-GCK SFAGKALDLCADGDPICS-DGRNPFAHTSYERSPLIGQAAGFAAGRL---
TH_4127|M.thermoresistible__bu VFGGRGIDLCSDGDPICS-NGRNPFAHTNYETSVHIETAADFAAGLI---
MSMEG_6354|M.smegmatis_MC2_155 LYTDKTVDLCIETDPVCSPTGSDGASHNLYATNGMVQQAADYVAARVLPQ
MAB_4405|M.abscessus_ATCC_1997 ELLHKVIDVCNPGDFFCDPQGGDIISHTTYAKDKLTDGAADTVVQRLVAL
: :::* * .*. * :* * . ** .. :
Mb3482|M.bovis_AF2122/97 -----------------------
Rv3452|M.tuberculosis_H37Rv -----------------------
MMAR_1097|M.marinum_M -----------------------
MUL_0855|M.ulcerans_Agy99 -----------------------
MAV_4396|M.avium_104 -----------------------
Mvan_1441|M.vanbaalenii_PYR-1 -----------------------
Mflv_4966|M.gilvum_PYR-GCK -----------------------
TH_4127|M.thermoresistible__bu -----------------------
MSMEG_6354|M.smegmatis_MC2_155 -----------------------
MAB_4405|M.abscessus_ATCC_1997 LGHRCKGADGQSTDSATTCSPTA