For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AVLALALSVGLSGCSGTEKGETSSAPSSRLLNFTAQTIDGADFAGSSLAGKKAVLWFWAPWCPTCQKEAP DLQKAATAHSDVTFVGVAAQDQVPAMRDFVTKYGLTFIQLADTDAKVWALYDVTHQPAFAFLGAEGKAEV VKSPLSGPELDKKIGQLH*
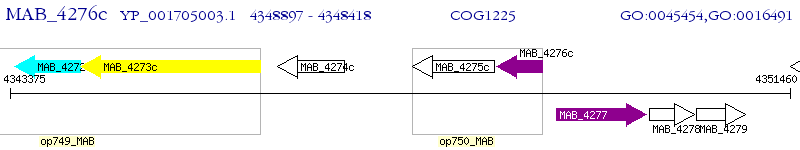
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4276c | - | - | 100% (159) | lipoprotein DsbF |
| M. abscessus ATCC 19977 | MAB_3243 | - | 1e-26 | 42.40% (125) | soluble secreted antigen MPT53 precursor |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1704 | dsbF | 1e-39 | 57.81% (128) | lipoprotein DsbF |
| M. gilvum PYR-GCK | Mflv_0052 | - | 3e-07 | 31.30% (115) | alkyl hydroperoxide reductase/ Thiol specific antioxidant/ Mal |
| M. tuberculosis H37Rv | Rv1677 | dsbF | 1e-39 | 57.81% (128) | lipoprotein DsbF |
| M. leprae Br4923 | MLBr_02703 | trx | 0.0001 | 27.27% (110) | bifunctional thioredoxin reductase/thioredoxin |
| M. marinum M | MMAR_5057 | dsbF | 1e-38 | 47.65% (170) | lipoprotein DsbF |
| M. avium 104 | MAV_3729 | - | 8e-33 | 46.31% (149) | 15 kDa antigen |
| M. smegmatis MC2 155 | MSMEG_3543 | - | 4e-34 | 49.28% (138) | soluble secreted antigen MPT53 |
| M. thermoresistible (build 8) | TH_2073 | mpt53 | 3e-34 | 43.48% (161) | SOLUBLE SECRETED ANTIGEN MPT53 PRECURSOR |
| M. ulcerans Agy99 | MUL_4133 | dsbF | 1e-35 | 44.63% (177) | lipoprotein DsbF |
| M. vanbaalenii PYR-1 | Mvan_0866 | - | 4e-07 | 31.30% (115) | redoxin domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1704|M.bovis_AF2122/97 ----------------------------MTHSRLIG--ALTVVAIIVTAC
Rv1677|M.tuberculosis_H37Rv ----------------------------MTHSRLIG--ALTVVAIIVTAC
MMAR_5057|M.marinum_M -----------------------------MPRRLIASLAGIAAALTIAAC
MUL_4133|M.ulcerans_Agy99 -----------------------------MPRRLIASLAGIAAALTIAAC
MAB_4276c|M.abscessus_ATCC_199 -------------------------------------MAVLALALSVGLS
MSMEG_3543|M.smegmatis_MC2_155 --------------------------------MKMGFLRTGATARPGVLG
TH_2073|M.thermoresistible__bu --------------------------------MKI---RISGTIRFLLLG
MAV_3729|M.avium_104 -------------------------------------MAATAFAVALLLG
Mflv_0052|M.gilvum_PYR-GCK ----------------------------MNRVVAFGRTLMAACAATAVFA
Mvan_0866|M.vanbaalenii_PYR-1 ----------------------------MSRVVALMRALFAACAATAVLA
MLBr_02703|M.leprae_Br4923 MNTTPSAHETIHEVIVIGSGPAGYTAALYAARAQLTPLVFEGTSFGGALM
Mb1704|M.bovis_AF2122/97 GSQPKSQPAVAPTG---------DAAAATQVPAGQT--VPAQLQFSAKTL
Rv1677|M.tuberculosis_H37Rv GSQPKSQPAVAPTG---------DAAAATQVPAGQT--VPAQLQFSAKTL
MMAR_5057|M.marinum_M GSSGTASPAG-------------APASSSNAVANRAAHVPAQLQFTTKTI
MUL_4133|M.ulcerans_Agy99 GSSGTASPAGGSSGTA---SPAGAPASSSNAVANRAAHVPAQLQFTTKTI
MAB_4276c|M.abscessus_ATCC_199 GCSGTE------------------KGETSSAPSSR------LLNFTAQTI
MSMEG_3543|M.smegmatis_MC2_155 TLSAVVATVVLATA---------LVLAP-SAVADDR------LQFTGTTL
TH_2073|M.thermoresistible__bu ---AVAA--ALTIA---------LAGAP-TAVADDR------LQFTGTTL
MAV_3729|M.avium_104 -----------------------LGDAPRAAATDDR------LQFTATTL
Mflv_0052|M.gilvum_PYR-GCK GCATGSDAVAQGGTFE---FVAPGGQTDIFYDPPQERGTPGPLSGPDLMD
Mvan_0866|M.vanbaalenii_PYR-1 GCATGSDAVAQGGTFE---FVAPGGQTDIFYDPPDKRGTPGPLSGPDLMD
MLBr_02703|M.leprae_Br4923 TTTEVENYPGFRNGITGPELMDDMREQALRFGAELRTEDVESVSLRGPIK
. :.
Mb1704|M.bovis_AF2122/97 DGHDFHGESLLGKPAVLWFWA------------------PWCPTCQGEAP
Rv1677|M.tuberculosis_H37Rv DGHDFHGESLLGKPAVLWFWA------------------PWCPTCQGEAP
MMAR_5057|M.marinum_M DGKPFSGESLAGKATVLWFWT------------------PWCPKCQQEAP
MUL_4133|M.ulcerans_Agy99 DGKPFSGESLAGKATVLWFST------------------PWCPKCQQEAP
MAB_4276c|M.abscessus_ATCC_199 DGADFAGSSLAGKKAVLWFWA------------------PWCPTCQKEAP
MSMEG_3543|M.smegmatis_MC2_155 SGASFDGSSLAGKPAVLWFWT------------------PWCPFCNAEAP
TH_2073|M.thermoresistible__bu SGTPFDGASLAGRPAVLWFWT------------------PWCPFCNAEAP
MAV_3729|M.avium_104 SGAPFNGASLQGKPAVLWFWT------------------PWCPYCNAEAP
Mflv_0052|M.gilvum_PYR-GCK TDRTISLDDFAGKVVVINVWG------------------QWCGPCRTEIT
Mvan_0866|M.vanbaalenii_PYR-1 TDRTISLDDFAGKVVVVNVWG------------------QWCGPCRTEIT
MLBr_02703|M.leprae_Br4923 SVVTAEGQTYQARAVILAMGTSVRYLQIPGEQELLGRGVSACATCDGSFF
.: .:: . * * .
Mb1704|M.bovis_AF2122/97 VVGQVAASH------PEVTFVGVAGLD-QVPAMQE---------------
Rv1677|M.tuberculosis_H37Rv VVGQVAASH------PEVTFVGVAGLD-QVPAMQE---------------
MMAR_5057|M.marinum_M IVAKAAAAN------PEATFVGVAARE-EASAMQA---------------
MUL_4133|M.ulcerans_Agy99 IVAKAAAAN------PEATFVGVAARE-EASAMQA---------------
MAB_4276c|M.abscessus_ATCC_199 DLQKAATAH------SDVTFVGVAAQD-QVPAMRD---------------
MSMEG_3543|M.smegmatis_MC2_155 SVGQVAAAN------PEVTFVGVAARS-DVAAMQG---------------
TH_2073|M.thermoresistible__bu NVSRVAAAN------PDVTFVGVAARA-DVAAMQD---------------
MAV_3729|M.avium_104 GVSRVAAAN------PGVTFVGVAAHS-DVGAMAN---------------
Mflv_0052|M.gilvum_PYR-GCK ELQKVYDATRD----RGVAFLGIDVRDNNIDAPRD---------------
Mvan_0866|M.vanbaalenii_PYR-1 QLQQVYQATRD----QGVAFLGIDVRDNNIDAPRD---------------
MLBr_02703|M.leprae_Br4923 RGQDIAVIGGGDSAMEEALFLTRFARSVTLVHRRDEFRASKIMLGRARNN
. *:
Mb1704|M.bovis_AF2122/97 ----FVNKYPVKTFTQLADTDGS-----------------VWANFG----
Rv1677|M.tuberculosis_H37Rv ----FVNKYPVKTFTQLADTDGS-----------------VWANFG----
MMAR_5057|M.marinum_M ----FVDKYQLGDVTQLADSDGQ-----------------VWSNFG----
MUL_4133|M.ulcerans_Agy99 ----FVDKYQLGDVTQLADSDGQ-----------------VWSNFG----
MAB_4276c|M.abscessus_ATCC_199 ----FVTKYGL-TFIQLADTDAK-----------------VWALYD----
MSMEG_3543|M.smegmatis_MC2_155 ----FVDKYNL-NFTNLNDADGS-----------------IWARYN----
TH_2073|M.thermoresistible__bu ----FVNTYNL-DFTNLNDADGS-----------------IWARYN----
MAV_3729|M.avium_104 ----FVSKYNL-NFTTLNDADGA-----------------IWARYG----
Mflv_0052|M.gilvum_PYR-GCK ----FIIDRAI-TFPSIYDPPMR-----------------TMIAFGGRYP
Mvan_0866|M.vanbaalenii_PYR-1 ----FIIDRGV-TFPSIYDPPMR-----------------TMIAFGGRYP
MLBr_02703|M.leprae_Br4923 DKIKFITNHTVVAVNGYTTVTGLRLRNTTTGEETTLVVTGVFVAIGHEPR
*: : . .
Mb1704|M.bovis_AF2122/97 VTQQPAYAFVDPHGNVDVVRG--------RMSQDELTRRVTALTSR----
Rv1677|M.tuberculosis_H37Rv VTQQPAYAFVDPHGNVDVVRG--------RMSQDELTRRVTALTSR----
MMAR_5057|M.marinum_M VTQQPAWAFISANGEVEVVKG--------GLSEAQLTERVNGLTGK----
MUL_4133|M.ulcerans_Agy99 VTQQPAWAFISANGEVEVVKG--------VLSEAQLTERVNGFTGK----
MAB_4276c|M.abscessus_ATCC_199 VTHQPAFAFLGAEGKAEVVKS--------PLSGPELDKKIGQLH------
MSMEG_3543|M.smegmatis_MC2_155 VPWQPAYVFYRADGSSTFVNNPT-----SAMSKQELSDRVAALNS-----
TH_2073|M.thermoresistible__bu VPWQPAYVFYRADGSSSFVNNPT-----AAMSEAELADRVAALKN-----
MAV_3729|M.avium_104 VPWQPAYVFYRADGSSTFVNNPT-----SAMPQDELAARVAALR------
Mflv_0052|M.gilvum_PYR-GCK TTVIPSTVVLDREHRVAAVFLR-------ELLAQDLQPVVERLAAEQ---
Mvan_0866|M.vanbaalenii_PYR-1 TTVIPSTVVLDREHRVAAVFLR-------ELLAEDLQPVVERLAAER---
MLBr_02703|M.leprae_Br4923 SSLVSDVVDIDPDGYVLVKGRTTSTSMDGVFAAGDLVDRTYRQAITAAGS
. . . . : :*
Mb1704|M.bovis_AF2122/97 --------------------------------------------------
Rv1677|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_5057|M.marinum_M --------------------------------------------------
MUL_4133|M.ulcerans_Agy99 --------------------------------------------------
MAB_4276c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_3543|M.smegmatis_MC2_155 --------------------------------------------------
TH_2073|M.thermoresistible__bu --------------------------------------------------
MAV_3729|M.avium_104 --------------------------------------------------
Mflv_0052|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0866|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_02703|M.leprae_Br4923 GCAAAIDAERWLAEHAGSKANETTEETGDVDSTDTTDWSTAMTDAKNAGV
Mb1704|M.bovis_AF2122/97 --------------------------------------------------
Rv1677|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_5057|M.marinum_M --------------------------------------------------
MUL_4133|M.ulcerans_Agy99 --------------------------------------------------
MAB_4276c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_3543|M.smegmatis_MC2_155 --------------------------------------------------
TH_2073|M.thermoresistible__bu --------------------------------------------------
MAV_3729|M.avium_104 --------------------------------------------------
Mflv_0052|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0866|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_02703|M.leprae_Br4923 TIEVTDASFFADVLSSNKPVLVDFWATWCGPCKMVAPVLEEIASEQRNQL
Mb1704|M.bovis_AF2122/97 --------------------------------------------------
Rv1677|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_5057|M.marinum_M --------------------------------------------------
MUL_4133|M.ulcerans_Agy99 --------------------------------------------------
MAB_4276c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_3543|M.smegmatis_MC2_155 --------------------------------------------------
TH_2073|M.thermoresistible__bu --------------------------------------------------
MAV_3729|M.avium_104 --------------------------------------------------
Mflv_0052|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0866|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_02703|M.leprae_Br4923 TVAKLDVDTNPEMAREFQVVSIPTMILFQGGQPVKRIVGAKGKAALLRDL
Mb1704|M.bovis_AF2122/97 --------
Rv1677|M.tuberculosis_H37Rv --------
MMAR_5057|M.marinum_M --------
MUL_4133|M.ulcerans_Agy99 --------
MAB_4276c|M.abscessus_ATCC_199 --------
MSMEG_3543|M.smegmatis_MC2_155 --------
TH_2073|M.thermoresistible__bu --------
MAV_3729|M.avium_104 --------
Mflv_0052|M.gilvum_PYR-GCK --------
Mvan_0866|M.vanbaalenii_PYR-1 --------
MLBr_02703|M.leprae_Br4923 SDVVPNLN