For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPRRLIASLAGIAAALTIAACGSSGTASPAGAPASSSNAVANRAAHVPAQLQFTTKTIDGKPFSGESLAG KATVLWFWTPWCPKCQQEAPIVAKAAAANPEATFVGVAAREEASAMQAFVDKYQLGDVTQLADSDGQVWS NFGVTQQPAWAFISANGEVEVVKGGLSEAQLTERVNGLTGK
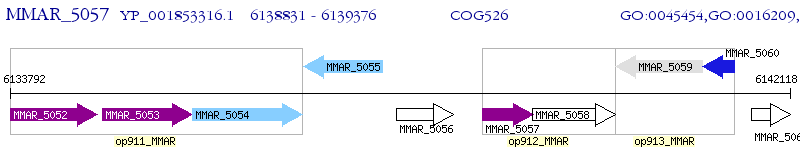
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_5057 | dsbF | - | 100% (181) | lipoprotein DsbF |
| M. marinum M | MMAR_1831 | mpt53 | 8e-34 | 47.18% (142) | soluble secreted antigen Mpt53 precursor |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1704 | dsbF | 4e-52 | 54.44% (180) | lipoprotein DsbF |
| M. gilvum PYR-GCK | Mflv_3216 | - | 4e-06 | 25.45% (165) | alkyl hydroperoxide reductase/ Thiol specific antioxidant/ Mal |
| M. tuberculosis H37Rv | Rv1677 | dsbF | 5e-52 | 54.44% (180) | lipoprotein DsbF |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4276c | - | 1e-38 | 47.65% (170) | lipoprotein DsbF |
| M. avium 104 | MAV_3729 | - | 1e-33 | 49.65% (141) | 15 kDa antigen |
| M. smegmatis MC2 155 | MSMEG_3543 | - | 2e-35 | 48.45% (161) | soluble secreted antigen MPT53 |
| M. thermoresistible (build 8) | TH_2073 | mpt53 | 2e-37 | 49.13% (173) | SOLUBLE SECRETED ANTIGEN MPT53 PRECURSOR |
| M. ulcerans Agy99 | MUL_4133 | dsbF | 7e-96 | 93.19% (191) | lipoprotein DsbF |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_5057|M.marinum_M -------MPRRLIASLAGIAAALTIAACGSSGTASPAG----------AP
MUL_4133|M.ulcerans_Agy99 -------MPRRLIASLAGIAAALTIAACGSSGTASPAGGSSGTASPAGAP
MSMEG_3543|M.smegmatis_MC2_155 --------MKMGFLRTGATARPGVLGTL------SAVV--------ATVV
TH_2073|M.thermoresistible__bu --------MKI---RISGTIRFLLLG---------AVA--------A--A
MAV_3729|M.avium_104 -------------MAATAFAVALLLG------------------------
Mb1704|M.bovis_AF2122/97 --------MTHSRLIGALTVVAIIVTACGSQPKSQPAV--------APTG
Rv1677|M.tuberculosis_H37Rv --------MTHSRLIGALTVVAIIVTACGSQPKSQPAV--------APTG
MAB_4276c|M.abscessus_ATCC_199 ---------------MAVLALALSVG---------------------LSG
Mflv_3216|M.gilvum_PYR-GCK MSETRSFTPHRWAGVTAVMLAAGALTACG------------------QSA
:
MMAR_5057|M.marinum_M ASSSNAVANRAAHVPAQLQFTTKTIDGKPFSGESLAGKATVLWFWTPWCP
MUL_4133|M.ulcerans_Agy99 ASSSNAVANRAAHVPAQLQFTTKTIDGKPFSGESLAGKATVLWFSTPWCP
MSMEG_3543|M.smegmatis_MC2_155 LATALVLAP-SAVADDRLQFTGTTLSGASFDGSSLAGKPAVLWFWTPWCP
TH_2073|M.thermoresistible__bu LTIALAGAP-TAVADDRLQFTGTTLSGTPFDGASLAGRPAVLWFWTPWCP
MAV_3729|M.avium_104 ----LGDAPRAAATDDRLQFTATTLSGAPFNGASLQGKPAVLWFWTPWCP
Mb1704|M.bovis_AF2122/97 DAAAATQVPAGQTVPAQLQFSAKTLDGHDFHGESLLGKPAVLWFWAPWCP
Rv1677|M.tuberculosis_H37Rv DAAAATQVPAGQTVPAQLQFSAKTLDGHDFHGESLLGKPAVLWFWAPWCP
MAB_4276c|M.abscessus_ATCC_199 CSGTEKGETSSAPSSRLLNFTAQTIDGADFAGSSLAGKKAVLWFWAPWCP
Mflv_3216|M.gilvum_PYR-GCK TTSNPTSPPHANATTQATAATLTTVDGKTVELP--AAAPTAILFFSYGCG
: *:.* . . :.: * : *
MMAR_5057|M.marinum_M KCQQEAPIVAKAAAANPEAT------FVGVAAREEASAMQAFVDKYQLGD
MUL_4133|M.ulcerans_Agy99 KCQQEAPIVAKAAAANPEAT------FVGVAAREEASAMQAFVDKYQLGD
MSMEG_3543|M.smegmatis_MC2_155 FCNAEAPSVGQVAAANPEVT------FVGVAARSDVAAMQGFVDKYNLN-
TH_2073|M.thermoresistible__bu FCNAEAPNVSRVAAANPDVT------FVGVAARADVAAMQDFVNTYNLD-
MAV_3729|M.avium_104 YCNAEAPGVSRVAAANPGVT------FVGVAAHSDVGAMANFVSKYNLN-
Mb1704|M.bovis_AF2122/97 TCQGEAPVVGQVAASHPEVT------FVGVAGLDQVPAMQEFVNKYPVKT
Rv1677|M.tuberculosis_H37Rv TCQGEAPVVGQVAASHPEVT------FVGVAGLDQVPAMQEFVNKYPVKT
MAB_4276c|M.abscessus_ATCC_199 TCQKEAPDLQKAATAHSDVT------FVGVAAQDQVPAMRDFVTKYGLT-
Mflv_3216|M.gilvum_PYR-GCK ECVGGGKSLAAARAAVEKAGGSAKFLAVDIVPTEKPADVRHFLDQIGGTS
* . : . :: . *.:. . : *:
MMAR_5057|M.marinum_M VTQLADSDGQVWSNFGVTQQPAWAFISANGEVEVVK---GGLSEAQLTER
MUL_4133|M.ulcerans_Agy99 VTQLADSDGQVWSNFGVTQQPAWAFISANGEVEVVK---GVLSEAQLTER
MSMEG_3543|M.smegmatis_MC2_155 FTNLNDADGSIWARYNVPWQPAYVFYRADGSSTFVNNPTSAMSKQELSDR
TH_2073|M.thermoresistible__bu FTNLNDADGSIWARYNVPWQPAYVFYRADGSSSFVNNPTAAMSEAELADR
MAV_3729|M.avium_104 FTTLNDADGAIWARYGVPWQPAYVFYRADGSSTFVNNPTSAMPQDELAAR
Mb1704|M.bovis_AF2122/97 FTQLADTDGSVWANFGVTQQPAYAFVDPHGNVDVVR---GRMSQDELTRR
Rv1677|M.tuberculosis_H37Rv FTQLADTDGSVWANFGVTQQPAYAFVDPHGNVDVVR---GRMSQDELTRR
MAB_4276c|M.abscessus_ATCC_199 FIQLADTDAKVWALYDVTHQPAFAFLGAEGKAEVVK---SPLSGPELDKK
Mflv_3216|M.gilvum_PYR-GCK LPAVVDTNGALTSRYQVTAPTTALVIDPSGQISYRG---HAPSQDQILAA
. : *::. : : : *. .: . . *. . ::
MMAR_5057|M.marinum_M VNGLTGK
MUL_4133|M.ulcerans_Agy99 VNGFTGK
MSMEG_3543|M.smegmatis_MC2_155 VAALNS-
TH_2073|M.thermoresistible__bu VAALKN-
MAV_3729|M.avium_104 VAALR--
Mb1704|M.bovis_AF2122/97 VTALTSR
Rv1677|M.tuberculosis_H37Rv VTALTSR
MAB_4276c|M.abscessus_ATCC_199 IGQLH--
Mflv_3216|M.gilvum_PYR-GCK LGSSAAR
: