For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NRVVAFGRTLMAACAATAVFAGCATGSDAVAQGGTFEFVAPGGQTDIFYDPPQERGTPGPLSGPDLMDTD RTISLDDFAGKVVVINVWGQWCGPCRTEITELQKVYDATRDRGVAFLGIDVRDNNIDAPRDFIIDRAITF PSIYDPPMRTMIAFGGRYPTTVIPSTVVLDREHRVAAVFLRELLAQDLQPVVERLAAEQ*
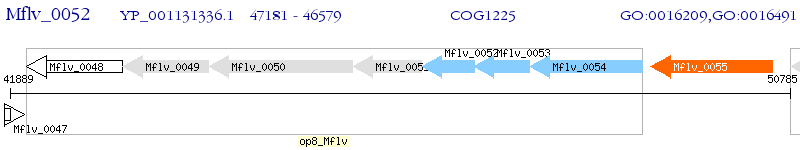
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0052 | - | - | 100% (200) | alkyl hydroperoxide reductase/ Thiol specific antioxidant/ Mal allergen |
| M. gilvum PYR-GCK | Mflv_2812 | - | 2e-83 | 76.60% (188) | alkyl hydroperoxide reductase/ Thiol specific antioxidant/ Mal |
| M. gilvum PYR-GCK | Mflv_2869 | - | 6e-64 | 60.10% (193) | alkyl hydroperoxide reductase/ Thiol specific antioxidant/ Mal |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0539 | - | 4e-82 | 72.40% (192) | thioredoxin protein |
| M. tuberculosis H37Rv | Rv0526 | - | 4e-82 | 72.40% (192) | thioredoxin protein |
| M. leprae Br4923 | MLBr_02412 | - | 2e-78 | 71.20% (191) | hypothetical protein MLBr_02412 |
| M. abscessus ATCC 19977 | MAB_3976c | - | 2e-74 | 67.19% (192) | thioredoxin |
| M. marinum M | MMAR_0872 | - | 8e-84 | 75.39% (191) | thioredoxin protein (thiol-disulfide interchange protein) |
| M. avium 104 | MAV_4619 | - | 4e-80 | 71.58% (190) | hypothetical protein MAV_4619 |
| M. smegmatis MC2 155 | MSMEG_0971 | - | 6e-86 | 76.50% (200) | hypothetical protein MSMEG_0971 |
| M. thermoresistible (build 8) | TH_2251 | - | 2e-92 | 86.89% (183) | POSSIBLE THIOREDOXIN PROTEIN (THIOL-DISULFIDE INTERCHANGE |
| M. ulcerans Agy99 | MUL_0625 | - | 2e-83 | 74.87% (191) | thioredoxin protein (thiol-disulfide interchange protein) |
| M. vanbaalenii PYR-1 | Mvan_0866 | - | 1e-106 | 91.50% (200) | redoxin domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0052|M.gilvum_PYR-GCK ----------MNR---VVAFGRTLMAACAATAVFAGCATGSDAVAQGGTF
Mvan_0866|M.vanbaalenii_PYR-1 ----------MSR---VVALMRALFAACAATAVLAGCATGSDAVAQGGTF
TH_2251|M.thermoresistible__bu ----------VKR---LVVLL-----VGTAMLVLTGCSTGDDAVAQGGTF
MSMEG_0971|M.smegmatis_MC2_155 ------MQSWVAR---VVGVTLAAL-FAVLTGALAGCSTGDDAVAQGGTF
MLBr_02412|M.leprae_Br4923 MIAPATMQSEAMRRSGATIIWRLANAGALLATLLAGCS-GRDAVVQGGTF
MAV_4619|M.avium_104 ---------MSAR--------VLAMAGAVLAGLLTGCSSGHDAVAQGGTF
Mb0539|M.bovis_AF2122/97 ------MQSRATRRSGALTMRRLVIAAAVSALLLTGCS-GRDAVAQGGTF
Rv0526|M.tuberculosis_H37Rv ------MQSRATRRSGALTMRRLVIAAAVSALLLTGCS-GRDAVAQGGTF
MMAR_0872|M.marinum_M -----------MR--------RLVIAAAVLAALLTGCS-GRDAVVQGGTF
MUL_0625|M.ulcerans_Agy99 ---------MTMR--------RLVIAAAVLAALLTGCS-GRDAVVQGGTF
MAB_3976c|M.abscessus_ATCC_199 -----------MR--------LLVAALAALALFITACSTGDDAVARGGQF
* ::.*: * ***.:** *
Mflv_0052|M.gilvum_PYR-GCK EFVAPGGQTDIFYDPPQERGTPGPLSGPDLMDTDRTISLDD--FAGKVVV
Mvan_0866|M.vanbaalenii_PYR-1 EFVAPGGQTDIFYDPPDKRGTPGPLSGPDLMDTDRTISLDD--FAGKVVV
TH_2251|M.thermoresistible__bu EFVAPGGQTDIFYDPPADRGRPGPISGPDLMNPDETISLDD--FAGQVVV
MSMEG_0971|M.smegmatis_MC2_155 EFVAPGGKTDIFYDPPQSRGRPGTLAGPLLTDPSKTISLDD--FAGEVVV
MLBr_02412|M.leprae_Br4923 EFVSPDGKTDIFYEPPASRGRPGPLSGLDLADPERTVALDD--FTGNVVV
MAV_4619|M.avium_104 EFVSPGGKTDIYYDPPSSRGRPGPLSGPDLADPAHTLSLDDPIFAGRVVV
Mb0539|M.bovis_AF2122/97 EFVSPGGKTDIFYDPPASRGRPGPLSGPELADPARSVSLDD--FPGQVVV
Rv0526|M.tuberculosis_H37Rv EFVSPGGKTDIFYDPPASRGRPGPLSGPELADPARSVSLDD--FPGQVVV
MMAR_0872|M.marinum_M EFVSPGGKTDIFYDPPASRGHPGPLSGPDLIDPDRTLSLDD--FAGRVVV
MUL_0625|M.ulcerans_Agy99 EFVSPGGKTDIFYDPPASRGHPGPLSGPDLIDPDRTLSLDD--FAGRVVV
MAB_3976c|M.abscessus_ATCC_199 QFVSPGGKTDILYDPPQSRQTPGPISGPSLLDPTKTLSLSD--FRGRVVV
:**:*.*:*** *:** .* **.::* * :. .:::*.* * *.***
Mflv_0052|M.gilvum_PYR-GCK INVWGQWCGPCRTEITELQKVYDATRDRGVAFLGIDVRDNNIDAPRDFII
Mvan_0866|M.vanbaalenii_PYR-1 VNVWGQWCGPCRTEITQLQQVYQATRDQGVAFLGIDVRDNNIDAPRDFII
TH_2251|M.thermoresistible__bu INIWGQWCGPCRTEILELQQVYDATRHQGVAFLGIDVRDNNIDAAQDFIA
MSMEG_0971|M.smegmatis_MC2_155 VNVWGQWCGPCRAEITQLQEVHNATKDKGVAFLGIDVRDNNRDAAVDFVK
MLBr_02412|M.leprae_Br4923 INVWGQWCGPCRTEVTQLQQVYNATRDSGVSFLGIDVRDNNRQAAQDFVN
MAV_4619|M.avium_104 INIWGQWCGPCRSEVSQLQQVYDATRGAGASFLGIDVRDNNRQAALDFVN
Mb0539|M.bovis_AF2122/97 VNVWGQWCGPCRAEVSQLQRVYDATRGAGVSFLGIDVRDNNRQAPQDFIN
Rv0526|M.tuberculosis_H37Rv VNVWGQWCGPCRAEVSQLQRVYDATRGAGVSFLGIDVRDNNRQAPQDFIN
MMAR_0872|M.marinum_M VNVWGQWCGPCRAEITQLQQVYDATRAAGVSFLGIDVRDNSRAAAQDFVR
MUL_0625|M.ulcerans_Agy99 VNVWGQWCGPCRAEITQLQQVYDETRAAGVSFLGIDVRDNSRAAAQDFVR
MAB_3976c|M.abscessus_ATCC_199 INVWGQWCGPCRSEFSALEDVYRATHAQGVEFLGINVRDNEITKAQDFVT
:*:*********:*. *: *: *: *. ****:****. . **:
Mflv_0052|M.gilvum_PYR-GCK DRAITFPSIYDPPMRTMIAFGGRYPTTVIPSTVVLDREHRVAAVFLRELL
Mvan_0866|M.vanbaalenii_PYR-1 DRGVTFPSIYDPPMRTMIAFGGRYPTTVIPSTVVLDREHRVAAVFLRELL
TH_2251|M.thermoresistible__bu DRNITFPSIYDPPMRTMIAFGGRYPTTVIPSTVVLDREHRVAAVFLRELL
MSMEG_0971|M.smegmatis_MC2_155 DRNVTFPSIYDPPMRTMIAFGGKYPTTVIPSTVVLDRQHRVAAVFLRELL
MLBr_02412|M.leprae_Br4923 DRQVTFPSIYDPAMRTLIAFGDKYPTTVIPFTLVLDRQHRVAAVFLRELL
MAV_4619|M.avium_104 DRHVTFPSIYDPAMRTLIAFGGKYPTTVIPSTLVLDRQHRVAAVFLRELL
Mb0539|M.bovis_AF2122/97 DRHVTYPSIYDPAMRTLIAFGGKYPTSVIPSTLVLDRQHRVAAVFLRELL
Rv0526|M.tuberculosis_H37Rv DRHVTYPSIYDPAMRTLIAFGGKYPTSVIPSTLVLDRQHRVAAVFLRELL
MMAR_0872|M.marinum_M DRHVTFPSIYDPAMRTLIAFGGKYPTTVIPSTLVLDRQHRVAAVFLRELL
MUL_0625|M.ulcerans_Agy99 DRHVTFPSIYDPAMRTLIAFGGKYPTTVIPSTLVLDRQHRVAAVFLRELL
MAB_3976c|M.abscessus_ATCC_199 DRKVPYPSIYDPSMRTLIAFGKKFPTGAIPATLVLDRQHRVAAVFLRELL
** :.:******.***:**** ::** .** *:****:************
Mflv_0052|M.gilvum_PYR-GCK AQDLQPVVERLAAEQ--------------
Mvan_0866|M.vanbaalenii_PYR-1 AEDLQPVVERLAAER--------------
TH_2251|M.thermoresistible__bu AEDLQPVVERLAAEDPAPDGDAS------
MSMEG_0971|M.smegmatis_MC2_155 AEDLEPLIERLLAEPAEPGEPAVSKEQRP
MLBr_02412|M.leprae_Br4923 AGDLQPVVARVAHETVAGQLPRGTS----
MAV_4619|M.avium_104 ATDLQPVVQRVAQQ---------------
Mb0539|M.bovis_AF2122/97 AADLQPVVERVAEEEPSGRAPVGAQ----
Rv0526|M.tuberculosis_H37Rv AADLQPVVERVAEEEPSGRAPVGAQ----
MMAR_0872|M.marinum_M AEDLQPVVQRLAQEPAAGSTPPGPQ----
MUL_0625|M.ulcerans_Agy99 AEDLQPVVQRLAQEPAAGSTPPGPQ----
MAB_3976c|M.abscessus_ATCC_199 AQDLRPVVERLAQESAKETAGEVK-----
* **.*:: *: :