For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTHSRLIGALTVVAIIVTACGSQPKSQPAVAPTGDAAAATQVPAGQTVPAQLQFSAKTLDGHDFHGESLL GKPAVLWFWAPWCPTCQGEAPVVGQVAASHPEVTFVGVAGLDQVPAMQEFVNKYPVKTFTQLADTDGSVW ANFGVTQQPAYAFVDPHGNVDVVRGRMSQDELTRRVTALTSR
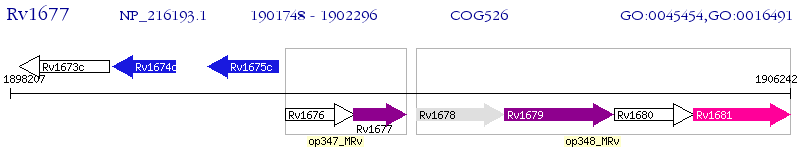
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1677 | dsbF | - | 100% (182) | PROBABLE CONSERVED LIPOPROTEIN DSBF |
| M. tuberculosis H37Rv | Rv2878c | mpt53 | 2e-35 | 52.82% (142) | soluble secreted antigen MPT53 precursor |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1704 | dsbF | 1e-105 | 100.00% (182) | lipoprotein DsbF |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4276c | - | 2e-39 | 57.81% (128) | lipoprotein DsbF |
| M. marinum M | MMAR_5057 | dsbF | 7e-52 | 54.44% (180) | lipoprotein DsbF |
| M. avium 104 | MAV_3729 | - | 6e-35 | 52.86% (140) | 15 kDa antigen |
| M. smegmatis MC2 155 | MSMEG_3543 | - | 7e-38 | 51.81% (166) | soluble secreted antigen MPT53 |
| M. thermoresistible (build 8) | TH_2073 | mpt53 | 2e-34 | 51.33% (150) | SOLUBLE SECRETED ANTIGEN MPT53 PRECURSOR |
| M. ulcerans Agy99 | MUL_4133 | dsbF | 3e-49 | 51.06% (188) | lipoprotein DsbF |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Rv1677|M.tuberculosis_H37Rv MTHSRLIG--ALTVVAIIVTACGSQPKSQPAVAPTG------DAAAATQV
Mb1704|M.bovis_AF2122/97 MTHSRLIG--ALTVVAIIVTACGSQPKSQPAVAPTG------DAAAATQV
MMAR_5057|M.marinum_M -MPRRLIASLAGIAAALTIAACGSSGTASPAG----------APASSSNA
MUL_4133|M.ulcerans_Agy99 -MPRRLIASLAGIAAALTIAACGSSGTASPAGGSSGTASPAGAPASSSNA
MAB_4276c|M.abscessus_ATCC_199 ---------MAVLALALSVGLSGCSGTE---------------KGETSSA
MSMEG_3543|M.smegmatis_MC2_155 ----MKMGFLRTGATARPGVLGTLSAVVATVVLATA------LVLAP-SA
TH_2073|M.thermoresistible__bu ----MKI---RISGTIRFLLLG---AVAA--ALTIA------LAGAP-TA
MAV_3729|M.avium_104 ---------MAATAFAVALLLG--------------------LGDAPRAA
. .
Rv1677|M.tuberculosis_H37Rv PAGQT--VPAQLQFSAKTLDGHDFHGESLLGKPAVLWFWAPWCPTCQGEA
Mb1704|M.bovis_AF2122/97 PAGQT--VPAQLQFSAKTLDGHDFHGESLLGKPAVLWFWAPWCPTCQGEA
MMAR_5057|M.marinum_M VANRAAHVPAQLQFTTKTIDGKPFSGESLAGKATVLWFWTPWCPKCQQEA
MUL_4133|M.ulcerans_Agy99 VANRAAHVPAQLQFTTKTIDGKPFSGESLAGKATVLWFSTPWCPKCQQEA
MAB_4276c|M.abscessus_ATCC_199 PSSR------LLNFTAQTIDGADFAGSSLAGKKAVLWFWAPWCPTCQKEA
MSMEG_3543|M.smegmatis_MC2_155 VADDR------LQFTGTTLSGASFDGSSLAGKPAVLWFWTPWCPFCNAEA
TH_2073|M.thermoresistible__bu VADDR------LQFTGTTLSGTPFDGASLAGRPAVLWFWTPWCPFCNAEA
MAV_3729|M.avium_104 ATDDR------LQFTATTLSGAPFNGASLQGKPAVLWFWTPWCPYCNAEA
:. *:*: *:.* * * ** *: :**** :**** *: **
Rv1677|M.tuberculosis_H37Rv PVVGQVAASHPEVTFVGVAGLDQVPAMQEFVNKYPVKTFTQLADTDGSVW
Mb1704|M.bovis_AF2122/97 PVVGQVAASHPEVTFVGVAGLDQVPAMQEFVNKYPVKTFTQLADTDGSVW
MMAR_5057|M.marinum_M PIVAKAAAANPEATFVGVAAREEASAMQAFVDKYQLGDVTQLADSDGQVW
MUL_4133|M.ulcerans_Agy99 PIVAKAAAANPEATFVGVAAREEASAMQAFVDKYQLGDVTQLADSDGQVW
MAB_4276c|M.abscessus_ATCC_199 PDLQKAATAHSDVTFVGVAAQDQVPAMRDFVTKYGL-TFIQLADTDAKVW
MSMEG_3543|M.smegmatis_MC2_155 PSVGQVAAANPEVTFVGVAARSDVAAMQGFVDKYNL-NFTNLNDADGSIW
TH_2073|M.thermoresistible__bu PNVSRVAAANPDVTFVGVAARADVAAMQDFVNTYNL-DFTNLNDADGSIW
MAV_3729|M.avium_104 PGVSRVAAANPGVTFVGVAAHSDVGAMANFVSKYNL-NFTTLNDADGAIW
* : :.*:::. .******. :. ** ** .* : . * *:*. :*
Rv1677|M.tuberculosis_H37Rv ANFGVTQQPAYAFVDPHGNVDVVRG---RMSQDELTRRVTALTSR
Mb1704|M.bovis_AF2122/97 ANFGVTQQPAYAFVDPHGNVDVVRG---RMSQDELTRRVTALTSR
MMAR_5057|M.marinum_M SNFGVTQQPAWAFISANGEVEVVKG---GLSEAQLTERVNGLTGK
MUL_4133|M.ulcerans_Agy99 SNFGVTQQPAWAFISANGEVEVVKG---VLSEAQLTERVNGFTGK
MAB_4276c|M.abscessus_ATCC_199 ALYDVTHQPAFAFLGAEGKAEVVKS---PLSGPELDKKIGQLH--
MSMEG_3543|M.smegmatis_MC2_155 ARYNVPWQPAYVFYRADGSSTFVNNPTSAMSKQELSDRVAALNS-
TH_2073|M.thermoresistible__bu ARYNVPWQPAYVFYRADGSSSFVNNPTAAMSEAELADRVAALKN-
MAV_3729|M.avium_104 ARYGVPWQPAYVFYRADGSSTFVNNPTSAMPQDELAARVAALR--
: :.*. ***:.* ..*. .*.. :. :* :: :