For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
EGFSGKVAVVTGAGSGIGRALAVELARSGAKIAISDIDNEGLAETERQIKSLGGDVRSDRLNVAEREAFL LYADAIKEHFGTVNQIYNNAGIDFHGELEASEFKDIERIIDVDFWGVVNGTKAFLPHLIASGDGHVVNVS SIFGVMACPGQSSYNAAKFAVRGFTESLRQEMIIGKKPVAVTCVHPGGIKTNVARNATVAEGYDHAEFTK LFDMLARTRPQAAARIILTAVRKKKPRVLVGPDAKAIDILVRLTGARYQDLFVLGERFLMPKRSH*
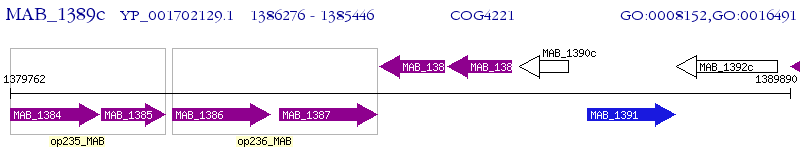
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1389c | - | - | 100% (276) | short-chain dehydrogenase/reductase |
| M. abscessus ATCC 19977 | MAB_1388c | - | e-129 | 82.85% (274) | short-chain dehydrogenase/reductase |
| M. abscessus ATCC 19977 | MAB_3919c | - | 3e-80 | 54.37% (263) | putative short chain dehydrogenase/reductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1277c | - | 1e-107 | 67.88% (274) | short-chain type dehydrogenase/reductase |
| M. gilvum PYR-GCK | Mflv_2216 | - | 1e-102 | 68.18% (264) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv1245c | - | 1e-107 | 67.88% (274) | short-chain type dehydrogenase/reductase |
| M. leprae Br4923 | MLBr_01094 | - | 1e-102 | 66.06% (277) | short chain alcohol dehydrogenase |
| M. marinum M | MMAR_4195 | - | 1e-106 | 71.32% (265) | short-chain type dehydrogenase/reductase |
| M. avium 104 | MAV_1384 | - | 1e-112 | 75.76% (264) | short chain alcohol dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_5053 | - | 1e-109 | 70.44% (274) | short chain alcohol dehydrogenase |
| M. thermoresistible (build 8) | TH_1828 | - | 1e-103 | 68.56% (264) | PROBABLE SHORT-CHAIN TYPE DEHYDROGENASE/REDUCTASE |
| M. ulcerans Agy99 | MUL_4501 | - | 1e-105 | 70.57% (265) | short-chain type dehydrogenase/reductase |
| M. vanbaalenii PYR-1 | Mvan_4479 | - | 1e-105 | 68.94% (264) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4195|M.marinum_M MEGFAGKVAVVTGAGSGIGRALAIELARSGAKLAISDVDTEGLAQTEKLV
MUL_4501|M.ulcerans_Agy99 MEGFAGKVAVVTGAGSGIGRALAIELARSGAKLAISDVDTEGLAQTEKLV
Mb1277c|M.bovis_AF2122/97 MEGFAGKVAVVTGAGSGIGQALAIELARSGAKVAISDVDTDGLADTEHRL
Rv1245c|M.tuberculosis_H37Rv MEGFAGKVAVVTGAGSGIGQALAIELARSGAKVAISDVDTDGLADTEHRL
Mflv_2216|M.gilvum_PYR-GCK MQGFAGKVAVVTGAGSGIGQALAIELGRSGALVAISDVDTEGLAVTEERL
Mvan_4479|M.vanbaalenii_PYR-1 MEGFAGKVAVVTGAGSGIGQALAIELGRSGARVAISDVDTEGLAVTEERL
TH_1828|M.thermoresistible__bu MQGFAGKVAVITGAGSGIGQALALELGRSGAKVAISDVNTEGLTETEDRL
MSMEG_5053|M.smegmatis_MC2_155 MEGFAGKVAVVTGAGSGIGQALAIELGRSGAKLAICDVDTEGLAATEERL
MLBr_01094|M.leprae_Br4923 MEGFAGKVAVVTGAGSGIGQALAIELARSGAKLAISDVDGEGLAQTEGQL
MAV_1384|M.avium_104 MQGFAGKVAVVTGAGSGIGQALAVELARSGAKVAISDVDLEGLAHTEEQL
MAB_1389c|M.abscessus_ATCC_199 MEGFSGKVAVVTGAGSGIGRALAVELARSGAKIAISDIDNEGLAETERQI
*:**:*****:********:***:**.**** :**.*:: :**: ** :
MMAR_4195|M.marinum_M TALGAEVKTDRLDVTEREAFLAYADAVNEHFGKVNQIYNNAGIGHTGDVE
MUL_4501|M.ulcerans_Agy99 TALGAEVKTDRLDVTERETFLAYADAVNEHFGKVNQIYNNAGIGHTGDVE
Mb1277c|M.bovis_AF2122/97 KAISTPVKTDRLDVTEREAFLAYADAVNEHFGTVNQIYNNAGIAFTGDIE
Rv1245c|M.tuberculosis_H37Rv KAISTPVKTDRLDVTEREAFLAYADAVNEHFGTVNQIYNNAGIAFTGDIE
Mflv_2216|M.gilvum_PYR-GCK KAIGTHVKSDRLNVTERERFLQYADDVAEHFGKVHQIYNNAGIAFTGDIE
Mvan_4479|M.vanbaalenii_PYR-1 KAIGAQVKSDRLDVTERETFLLYADDVAEHFGSVNQIYNNAGIAFTGDIE
TH_1828|M.thermoresistible__bu KAIGAPVKADRLDVTERAAFEAYADAVVEHFGVVNQIYNNAGIAYTGDIE
MSMEG_5053|M.smegmatis_MC2_155 KAIGAPVKADRLDVTEREAFLLYADAVKEHYGTVNQIYNNAGIAFTGDVE
MLBr_01094|M.leprae_Br4923 KAIGASARTDRLDVTEREAFLTYADVVHENFGKVNQIYNNAGIAFTGDVE
MAV_1384|M.avium_104 KAIGAQYKADRLDVTEREAFLAYADAVKEHFGKVNQIYNNAGIAFTGDVE
MAB_1389c|M.abscessus_ATCC_199 KSLGGDVRSDRLNVAEREAFLLYADAIKEHFGTVNQIYNNAGIDFHGELE
.::. ::***:*:** * *** : *::* *:******** . *::*
MMAR_4195|M.marinum_M VCAFKDIDRVMDVDFGGVLNGTKAFLPYLIASGDGHVINVSSVFGLFSVS
MUL_4501|M.ulcerans_Agy99 VCAFKDIDRVMDVDFGGVLNGTKTFLPYLIASGDGHVINVSSVFGLFSVS
Mb1277c|M.bovis_AF2122/97 VSQFKDIERVMDVDFWGVVNGTKAFLPHLIASGDGHVINISSVFGLFSAP
Rv1245c|M.tuberculosis_H37Rv VSQFKDIERVMDVDFWGVVNGTKAFLPHLIASGDGHVINISSVFGLFSAP
Mflv_2216|M.gilvum_PYR-GCK VSQFKDIERVMDVDFWGVVNGTKAFLPHLIASGDGHVVNVSSVFGLFSVP
Mvan_4479|M.vanbaalenii_PYR-1 ITQFKDIERVMDVDYWGVVNGTKAFLPHLIASGDGHVVNVSSVFGLFSVP
TH_1828|M.thermoresistible__bu VSHFKDIERVMDVDFWGVVNGTKVFLPHLIASGDGHVINISSVFGLFSVP
MSMEG_5053|M.smegmatis_MC2_155 ISEFKDIERVMDVDYWGVVNGTKAFLPHLIASGDGHVINVSSVFGLFSVP
MLBr_01094|M.leprae_Br4923 VSHFKDIERVMDVDYWGVVNGTKAFLPYLISSGDGHVINISSVFGLFSVP
MAV_1384|M.avium_104 VSQFKDIERVMDVDFWGVVNGTKAFLPHLIASGDGHVVNVSSVFGLFSVP
MAB_1389c|M.abscessus_ATCC_199 ASEFKDIERIIDVDFWGVVNGTKAFLPHLIASGDGHVVNVSSIFGVMACP
****:*::***: **:****.***:**:******:*:**:**::: .
MMAR_4195|M.marinum_M GQAAYNAAKFAVRGFTEALRQEMILAGHPVGVTTVHPGGIKTAIARNATA
MUL_4501|M.ulcerans_Agy99 GQAAYNAAKFAVRGFTEALRQEMILAGHPVGVTTVHPGGIKTAIARNATA
Mb1277c|M.bovis_AF2122/97 GQAAYNSAKFAVRGFTEALRQEMALAGHPVKVTTVHPGGVKTAIARNATA
Rv1245c|M.tuberculosis_H37Rv GQAAYNSAKFAVRGFTEALRQEMALAGHPVKVTTVHPGGVKTAIARNATA
Mflv_2216|M.gilvum_PYR-GCK GQGAYNAAKFAVRGFTEALRQEMALAGHPVKVSCVHPGGIKTAIARNAGA
Mvan_4479|M.vanbaalenii_PYR-1 GQAAYNSAKFAVRGFTEALRQEMALAGHPVKVSCVHPGGIKTAIARNASA
TH_1828|M.thermoresistible__bu GQAAYNAAKFAVRGFTEALRQEMILAGHPVKVTTVHPGGIKTGIARNMTA
MSMEG_5053|M.smegmatis_MC2_155 GQAAYNSAKFAVRGFTEALRQEMAIAKHPVKVTTVHPGGIKTAIARNATA
MLBr_01094|M.leprae_Br4923 GQAAYNSAKFAVRGFTEALREEMALAGRPVNVTTVYPGGIKTAIARNATA
MAV_1384|M.avium_104 GQAAYNSAKFAVRGFTEALRQEMAAAGHPVAVTTVHPGGIKTAIARNATA
MAB_1389c|M.abscessus_ATCC_199 GQSSYNAAKFAVRGFTESLRQEMIIGKKPVAVTCVHPGGIKTNVARNATV
**.:**:**********:**:** . :** *: *:***:** :*** .
MMAR_4195|M.marinum_M AEGLDSDELAKMFDKRVARTSPERAAKIILGAVRKNKARVLVGPDAKALD
MUL_4501|M.ulcerans_Agy99 AEGLDSDELAKMFDKRVARTSPERAAKIILGAVRKNKARVLVGPDAKALD
Mb1277c|M.bovis_AF2122/97 AEGLDQAELAETFDKRVAHLSPQRAAQIILTGVAKNKARVLVGVDAKVLD
Rv1245c|M.tuberculosis_H37Rv AEGLDQAELAETFDKRVAHLSPQRAAQIILTGVAKNKARVLVGVDAKVLD
Mflv_2216|M.gilvum_PYR-GCK AEGLDAEELAKTFDKKLASTTPEKAAQVILDGVRKNKARILIGNDAKMFE
Mvan_4479|M.vanbaalenii_PYR-1 AEGLDAEELAKAFDKKLASTTPEKAAKIILDGVRKNRARILVGNDAKVFD
TH_1828|M.thermoresistible__bu AEGLDAQELARAFDTKLARTSPEKAAQIILDGVRKNKARVLVGTDAKILD
MSMEG_5053|M.smegmatis_MC2_155 AEGLDAKELAEAFDKKLANTTPQRAAVIILDGVRKNKARVLVGPDAKILD
MLBr_01094|M.leprae_Br4923 AEGLDVSKIASRFDTWVAHTSPQHAARIILKAVRKKKARVLVGPDAKVAN
MAV_1384|M.avium_104 AEGLDQAELAKLFDKRLAKTTPQRAAQIILDAVRKKKARVLVGSDAKALD
MAB_1389c|M.abscessus_ATCC_199 AEGYDHAEFTKLFD-MLARTRPQAAARIILTAVRKKKPRVLVGPDAKAID
*** * ::: ** :* *: ** :** .* *::.*:*:* *** :
MMAR_4195|M.marinum_M IVVRLTG-SGYQRLFMPVLGRLVPASHR
MUL_4501|M.ulcerans_Agy99 IVVRLTG-SGYQRLFMPVLGRLVPASHR
Mb1277c|M.bovis_AF2122/97 LVVRLTG-SGYQRIFPIITGRLIPRPR-
Rv1245c|M.tuberculosis_H37Rv LVVRLTG-SGYQRIFPIITGRLIPRPR-
Mflv_2216|M.gilvum_PYR-GCK IAIRALG-AGYQSVFPKVVARLTPPAK-
Mvan_4479|M.vanbaalenii_PYR-1 VVVRVLG-AGYQSLFPKAVARLTPPAK-
TH_1828|M.thermoresistible__bu AIVRLTG-SGYQRLFPKFVARTMPR---
MSMEG_5053|M.smegmatis_MC2_155 VIVRITG-SGYQRLFSAVAAKAMPR---
MLBr_01094|M.leprae_Br4923 VVVRFSGGAGYQRLFAQVASRLILNQR-
MAV_1384|M.avium_104 ILVRLTG-SGYQRLFGPVMSRLLPN---
MAB_1389c|M.abscessus_ATCC_199 ILVRLTG-ARYQDLFVLGERFLMPKRSH
:* * : ** :*