For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GAGSAGSILAGRLSEDPSFRVVLVEAGPVSSADEAGVRNGYRLPIGPGSQIATYYWATLTSTGDQAELVR GSVVGGSGAINGGYFVRGRPADFDGWAVSGWSWAEVRDHFRAIETDHDFRDDPLHGSQGPIPVGRRHEQS SAGTELMDLAVNRGYPVVADLNGSAGAGVGLVPLNIDSGIRVGPARAYLESAVWRPNLAVYSGTRVLRVL CSGGKATGVQVAVGNSVRTLTADRIILSAGAIESAKLLLLSGLGPADELRAVGVQPVVDLPGVGTRVMDH AEWVLDTGEQGQQGHPVLDTVLHTDHPMCLEIRPYTTGFAAMAGVVADGADARQIGVALMTPQCRGRLEL RSTDPAAPVYIDLRYDSETADMDRLRDGVHLVSELYGRRFGGPRWSTSQHLCGTAPLGNDRDKHAVVDKY CRVRGIDGLFVIDGSILPTIPSRGPAATIAMIGHRAAQFVAWS*
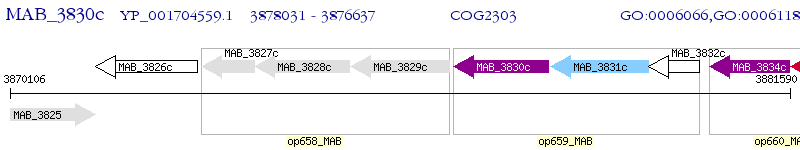
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3830c | - | - | 100% (464) | dehydrogenase (glucose-methanol-choline oxidoreductase) |
| M. abscessus ATCC 19977 | MAB_0250 | - | 3e-34 | 28.81% (531) | GMC-type oxidoreductase |
| M. abscessus ATCC 19977 | MAB_0816 | - | 1e-32 | 27.68% (513) | glucose-methanol-choline oxidoreductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0716 | - | 1e-116 | 51.17% (471) | dehydrogenase |
| M. gilvum PYR-GCK | Mflv_5056 | - | 1e-117 | 51.48% (474) | glucose-methanol-choline oxidoreductase |
| M. tuberculosis H37Rv | Rv0697 | - | 1e-116 | 51.17% (471) | dehydrogenase |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_1025 | - | 1e-118 | 52.55% (470) | dehydrogenase |
| M. avium 104 | MAV_4474 | - | 1e-117 | 53.18% (472) | FAD dependent oxidoreductase, putative |
| M. smegmatis MC2 155 | MSMEG_1428 | - | 1e-120 | 52.64% (473) | glucose-methanol-choline oxidoreductase |
| M. thermoresistible (build 8) | TH_4588 | - | 4e-70 | 50.83% (301) | PUTATIVE glucose-methanol-choline oxidoreductase |
| M. ulcerans Agy99 | MUL_0777 | - | 1e-118 | 52.55% (470) | dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_1300 | - | 1e-113 | 50.21% (472) | glucose-methanol-choline oxidoreductase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0716|M.bovis_AF2122/97 ----MTAAVRHSDVLVVGAGSAGSVVAERLSMDSSCVVTVLEAGPGLADP
Rv0697|M.tuberculosis_H37Rv ----MTAAVRHSDVLVVGAGSAGSVVAERLSMDSSCVVTVLEAGPGLADP
MAV_4474|M.avium_104 ----MNDAVSHSDVLIVGAGSAGSVVAERLSADPGCTVTVVEAGPSLDDP
MMAR_1025|M.marinum_M ----MTAALRHSDVLIVGAGSAGSVLAARLSADPSCAVTLVEAGPGLADP
MUL_0777|M.ulcerans_Agy99 ----MTAALRHSDVLVVGAGSAGSVLAARLSTDPSCAVTLVEAGPGLADP
MAB_3830c|M.abscessus_ATCC_199 ----------------MGAGSAGSILAGRLSEDPSFRVVLVEAGP--VSS
MSMEG_1428|M.smegmatis_MC2_155 -------------MLIVGAGSAGSVLAERLSADPSCQVTVVEAGPAPSDP
Mflv_5056|M.gilvum_PYR-GCK -----------MSVLIVGAGSAGSVLAERLSADPGCEVTVVEAGPGLSDS
Mvan_1300|M.vanbaalenii_PYR-1 ---------MRSDVLIVGAGSAGSVLAERLSADDGCQVTVVEAGPGPFDP
TH_4588|M.thermoresistible__bu VTGRPGAERCHSDVLIVGAGSAGSVLAERLSADSGCRVTVVEAGPGPSDP
:*******::* *** * . *.::**** ..
Mb0716|M.bovis_AF2122/97 GLLAQTANGLQLPIGAGSPLVERYRTRLTDRPVRHLPIVRGATVGGSGAI
Rv0697|M.tuberculosis_H37Rv GLLAQTANGLQLPIGAGSPLVERYRTRLTDRPVRHLPIVRGATVGGSGAI
MAV_4474|M.avium_104 ALLARTANGLQLPIGVGSPLVRRYQTQLTERPARRHPLVRGATIGGSGAI
MMAR_1025|M.marinum_M RLLAQTANGLQLPISADSPLVQRYQSQLTDHPVRRIPILRGATLGGSGAV
MUL_0777|M.ulcerans_Agy99 RLLAQTANGLQLPISADSPLVQCYQSQLTDHPVRRTPILRGATLGGSGAV
MAB_3830c|M.abscessus_ATCC_199 ADEAGVRNGYRLPIGPGSQIATYYWATLTS-TGDQAELVRGSVVGGSGAI
MSMEG_1428|M.smegmatis_MC2_155 RVAAQITDGVRLPIGAASSVVRHYGSTLTEDPPRRTEIIRGSVVGGSGAV
Mflv_5056|M.gilvum_PYR-GCK RVRDQISDGLRLPIGAASSVVRRYDTALTEEPRRHARIMRGVVVGGSGAV
Mvan_1300|M.vanbaalenii_PYR-1 RVGTQITDGLRLPIGVASSVVRRYPTALTDEPRRHAQIMRGAVVGGSGAV
TH_4588|M.thermoresistible__bu RVAHQITDGLRLPIGDASSVVRRYPTTLTDHPRRPAQLMRGAVVGGSGAV
:* :***. * :. * : **. . ::** .:*****:
Mb0716|M.bovis_AF2122/97 NGGYFCRGLPSDFDRASIPGWAWSDVLEHFRAIETDLDF-ETPVHGRSGP
Rv0697|M.tuberculosis_H37Rv NGGYFCRGLPSDFDRASIPGWAWSDVLEHFRAIETDLDF-ETPVHGRSGP
MAV_4474|M.avium_104 NGGYFCRGLRRDFDRLGLPGWAWSDVLDSFRAIETDLDF-DGPAHGRSGP
MMAR_1025|M.marinum_M NGGYFCRGLARDFDGAAIPGWSWSQVLPHFRAIETDLDF-DTPVHGRTGP
MUL_0777|M.ulcerans_Agy99 NGGYFCRGLARDFDGAAIPGWSWSQVLPHFRAIETDLDF-DTPVHGRTGP
MAB_3830c|M.abscessus_ATCC_199 NGGYFVRGRPADFDGWAVSGWSWAEVRDHFRAIETDHDFRDDPLHGSQGP
MSMEG_1428|M.smegmatis_MC2_155 NGGYFCRGLPTDFTAWAVPGWSWDDVLPHFRAIETDLDF-ATALHGSEGP
Mflv_5056|M.gilvum_PYR-GCK NGGYFCRALPADVDGWGMDGWSWDDVLPHFRAIETDLDF-ATPVHGDTGP
Mvan_1300|M.vanbaalenii_PYR-1 NGGYFCRALPSDFDGWGIAGWSWDEVLPHYTAIETDLDF-TGPLHGSAGP
TH_4588|M.thermoresistible__bu NGGYFCRGLPADFDRWGLGDWTWDRVLPHFRAIETDLDF-DGPLHGADGP
***** *. *. .: .*:* * : ***** ** . ** **
Mb0716|M.bovis_AF2122/97 IPVRRTHEMTGITESFMAAAEDAGFAWIADLN--DVGPEMPSGVGAVPLN
Rv0697|M.tuberculosis_H37Rv IPVRRTHEMTGITESFMAAAEDAGFAWIADLN--DVGPEMPSGVGAVPLN
MAV_4474|M.avium_104 ILVRRTREMTGTTERFIDAAWRAGFGWIDDLN--DAAAQPVSGIGAVPLN
MMAR_1025|M.marinum_M IPVRRTRDQTGVTQRFVTAAQRAGFGWIADLN--DVGPDIAAGVGAVPLN
MUL_0777|M.ulcerans_Agy99 IPVRRIRDQTGVTQRFVTAAQRAGFGWIADLN--DVGPDIAAGVGAVPLN
MAB_3830c|M.abscessus_ATCC_199 IPVGRRHEQSSAGTELMDLAVNRGYPVVADLN--GSAG---AGVGLVPLN
MSMEG_1428|M.smegmatis_MC2_155 IKVRRVHEFDGCTAAFVDAVSAAGYGWIDDLNGSDADAALPPGVGAVPLN
Mflv_5056|M.gilvum_PYR-GCK ITVRRVAEFDGATASFVDAARTAGFGWVEDLNGATAEHAVPAGIGAVPLN
Mvan_1300|M.vanbaalenii_PYR-1 ISVRRVSEFDGCTASFVESAREAGFGWIEDLNGSTPERPLPAGVGAVPLN
TH_4588|M.thermoresistible__bu IPVRRSTEFDGCTAKFIEAVEDAGYPWIPDLNGATPDQPLPPGVGQVPLN
* * * : . :: . *: : *** .*:* ****
Mb0716|M.bovis_AF2122/97 IVNGVRTSSAVGYLMPALGRPNLTLLARTRAVRLRFSATTAVGVDAIGPG
Rv0697|M.tuberculosis_H37Rv IVNGVRTSSAVGYLMPALGRPNLTLLARTRAVRLRFSATTAVGVDAIGPG
MAV_4474|M.avium_104 IVDGVRKGSGAGYLIPALSRPNLRVQAHTRVARLRFAGTVARGADVVGPH
MMAR_1025|M.marinum_M IVDGVRTGSGAAYLLPALGRPNLTLLEETRAQQVRFSAATAVGVDAVGPR
MUL_0777|M.ulcerans_Agy99 IVDGVRTGSGAAYLLPALGRPNLTLLEETRAQQVRFSAATAVGVDAVGPR
MAB_3830c|M.abscessus_ATCC_199 IDSGIRVGPARAYLESAVWRPNLAVYSGTRVLRVLCSGGKATGVQVAVGN
MSMEG_1428|M.smegmatis_MC2_155 IDGGTRLGPGGAYLQPALDRPNLHLRADTRVRRVLVEHDAAVGVECVDG-
Mflv_5056|M.gilvum_PYR-GCK IDRGTRVGPGGAFLQPALGRANLTVLTDTRVLRLTFAAGRVTGVLCVGPD
Mvan_1300|M.vanbaalenii_PYR-1 INGGTRVGPGGAFLQPALQRANLTLVTDTRVGGLRFSAGRVVGVDCLGPG
TH_4588|M.thermoresistible__bu INGGTRVGPGAAYLQPALARPNLTVLTETRVRRVRIADGRADGVECLGPD
* * * ... .:* .*: *.** : **. : . *.
Mb0716|M.bovis_AF2122/97 GPVSLSADRIVLCAGAIQSAHLLMLSGVGEEEVLRSAGVKVLMALP-VGM
Rv0697|M.tuberculosis_H37Rv GPVSLSADRIVLCAGAIQSAHLLMLSGVGEEEVLRSAGVKVLMALP-VGM
MAV_4474|M.avium_104 GRATLTADRIVLCAGAIETAHLLMLSGIGEPGMLRAAGVPVRVALP-VGT
MMAR_1025|M.marinum_M GPMTLWADRIVLCAGAIGSAHLLMLSGIGDHAMLRSAGVPVVQALP-VGM
MUL_0777|M.ulcerans_Agy99 GPMTLWADRIVLCAGAIGSAHLLMLSGIGDHAMLRAAGVPVVQALP-VGM
MAB_3830c|M.abscessus_ATCC_199 SVRTLTADRIILSAGAIESAKLLLLSGLGPADELRAVGVQPVVDLPGVGT
MSMEG_1428|M.smegmatis_MC2_155 --EILYADRIVLSAGAIGSAHLLLLSGIGPAGDLAAHGIAVAANLP-VGT
Mflv_5056|M.gilvum_PYR-GCK GPVTVGADRIVLCAGAIATAQLLMVSGVGPPPVLRAAGVPVEIDLP-VGV
Mvan_1300|M.vanbaalenii_PYR-1 GPTTLVADRVVLCAGAIASAQLLMVSGVGPQQVLRAAGVAVQIDLP-VGV
TH_4588|M.thermoresistible__bu GATVLTADRIVLSAGAIGSAHLLMLSGIGPQRLLGALGIPVVAAXXXXGV
: ***::*.**** :*:**::**:* * : *: *
Mb0716|M.bovis_AF2122/97 GCSDHPEWVMPTNWAVAVDRPVLEVLLSTHD--GIEIRPYTGGFVAMTGD
Rv0697|M.tuberculosis_H37Rv GCSDHPEWVMPTNWAVAVDRPVLEVLLSTHD--GIEIRPYTGGFVAMTGD
MAV_4474|M.avium_104 SFSDHAEWVLPTDWPVAPGRPVLEVVLSTAD--DLEIRPYTGGFVAMTGD
MMAR_1025|M.marinum_M NCSDHPEWALPTTWTVAAGRPVLEAVLNTAD--GIEIRPYTTGFAAMAGA
MUL_0777|M.ulcerans_Agy99 NCSDHPEWALPTTWTVVAGRPVLEAVLNTAD--GIEIRPYTTGFEAMAGA
MAB_3830c|M.abscessus_ATCC_199 RVMDHAEWVLDTGEQGQQGHPVLDTVLHTDHPMCLEIRPYTTGFAAMAGV
MSMEG_1428|M.smegmatis_MC2_155 ATVDHPEWVLPVAWTPTHDLPPLEAVLTTAD--GIEIRPYTAGFSALVHG
Mflv_5056|M.gilvum_PYR-GCK DTIDHPEWVLPVSWAPTHGVPPLEAVLTTDD--GVEIRPYTVGFEAMMTG
Mvan_1300|M.vanbaalenii_PYR-1 DTVDHPEWVLPVTWPPTHGVPPLEAVLTTGD--GLEIRPYTAGFGAMMTG
TH_4588|M.thermoresistible__bu GEPDPAHLAGQVDGGQGVDRDARRVTLHREHR-GPRIG---GGQHVEHRR
* .. . . . . * . .* * .
Mb0716|M.bovis_AF2122/97 GTAGHRDWPHIGVALMQPRARG--RITLVSSDPQIPVRIEHRYDSEPA-D
Rv0697|M.tuberculosis_H37Rv GTAGHRDWPHIGVALMQPRARG--RITLVSSDPQIPVRIEHRYDSEPA-D
MAV_4474|M.avium_104 GTAGHADWPHLGVALMQPRARG--CLTLMSADPAVPPRIEHRYDSEPG-D
MMAR_1025|M.marinum_M AAKGHRDWPHLGVALMRPRSRA--RVTLASPDPSVPPRIGHHYDSEPA-D
MUL_0777|M.ulcerans_Agy99 AAKGHRDWPHLGVALMRPRSRA--RVTLASPDPSVPPRIGHHYDSEPA-D
MAB_3830c|M.abscessus_ATCC_199 VADG-ADARQIGVALMTPQCRG--RLELRSTDPAAPVYIDLRYDSETA-D
MSMEG_1428|M.smegmatis_MC2_155 PEHDPAELPHLGVALMRPHSRG--RVRLASADPAVPPIIEHRYDTVAG-D
Mflv_5056|M.gilvum_PYR-GCK RRDDPTDQPHLGVTLMRPLSRG--RVTICSADPDADPVIEHRYDSAPA-D
Mvan_1300|M.vanbaalenii_PYR-1 RRDDPTDQPHLGVTLMQPLSRG--RVTITSADPDADPVIEHRYDSEPA-D
TH_4588|M.thermoresistible__bu GRRVEHERLLAGEPPLVARAAGGGRRHVGDQRRGRALGIGHGGDELPGGD
: * . : . . . : . * * .. *
Mb0716|M.bovis_AF2122/97 VAALRQGSALAHELCGAATRIGPAVWATSQHLCGSAPMGTDDDPRAVVDP
Rv0697|M.tuberculosis_H37Rv VAALRQGSALAHELCGAATRIGPAVWATSQHLCGSAPMGTDDDPRAVVDP
MAV_4474|M.avium_104 VAALARGAELARELAGGTTDAAAAIWSTSQHLCGTAPMGVDGDPRAVVDP
MMAR_1025|M.marinum_M VAALQRGTELARELAGSAVAGEPG-WSTSQHLCGSAPMGVDADPTAVVDP
MUL_0777|M.ulcerans_Agy99 VAALQRGTELARELAGSAVAGEPG-WSTSQHLCGSAPMGVDADPTAVVDP
MAB_3830c|M.abscessus_ATCC_199 MDRLRDGVHLVSELYG--RRFGGPRWSTSQHLCGTAPLGNDRDKHAVVDK
MSMEG_1428|M.smegmatis_MC2_155 VDALRAGAELARELVSHAVKVGEASWSTSQHLAGTAPLSADG--QGVLDP
Mflv_5056|M.gilvum_PYR-GCK VALLREGAQLASELAGATVQSEPS-WATSQHLCGTAKMG------AVVDE
Mvan_1300|M.vanbaalenii_PYR-1 VELLRAGTRLAHELAGATVQSEAS-WATSQHLCGTARMG------AVVDA
TH_4588|M.thermoresistible__bu PRQHRRPFVTGAELVEQVRHHDRAGQIRPAQQRAAHLLGDDAQLDRSRAR
** . : .: :.
Mb0716|M.bovis_AF2122/97 RCRVRG-IENLWVIDGSVLP---SITSRGPHATIVMLGHRAAEFVQ----
Rv0697|M.tuberculosis_H37Rv RCRVRG-IENLWVIDGSVLP---SITSRGPHATIVMLGHRAAEFVQ----
MAV_4474|M.avium_104 RCRVRG-IDNLWVIDGSVLP---AITSRGPHATIVMIGHRAAEFVR----
MMAR_1025|M.marinum_M ECRVRG-IENLWVVDGSILP---LIPSRGPHATIVMLGHRAAEFIG----
MUL_0777|M.ulcerans_Agy99 ECRVRG-IENLWVVDGSILP---LIPSRGPHATIVMLGHRAAEFIG----
MAB_3830c|M.abscessus_ATCC_199 YCRVRG-IDGLFVIDGSILP---TIPSRGPAATIAMIGHRAAQFVAWS--
MSMEG_1428|M.smegmatis_MC2_155 TCRVLG-VERLWVVDGSIMP---AITSRGPHATIAMIGHRAAEFIAT---
Mflv_5056|M.gilvum_PYR-GCK RCRVYG-VDNLWVVDGSILP---DIPSRGPHATIVMAGHRAAEFVSSS--
Mvan_1300|M.vanbaalenii_PYR-1 RCRVYG-VSNLWVVDGSILP---AIPSRGPHATIVMAGHRAAEFVSATST
TH_4588|M.thermoresistible__bu SAEVLGNHQAEQTHPGQLLPQRRIVAGVGRHQRPHLLLRRALLHEVGDSF
..* * . . *.::* :.. * : :** .
Mb0716|M.bovis_AF2122/97 -----
Rv0697|M.tuberculosis_H37Rv -----
MAV_4474|M.avium_104 -----
MMAR_1025|M.marinum_M -----
MUL_0777|M.ulcerans_Agy99 -----
MAB_3830c|M.abscessus_ATCC_199 -----
MSMEG_1428|M.smegmatis_MC2_155 -----
Mflv_5056|M.gilvum_PYR-GCK --AGA
Mvan_1300|M.vanbaalenii_PYR-1 ERAGA
TH_4588|M.thermoresistible__bu A----