For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LSAADVGRTISRIAHQIIEKTALSDSGDAPRVVLVGIPTRGATLAKRLAAHITEFSGVEVPAGFLDITLY RDDLRNKPHRPLERTSIPEGGVDGALVVLVDDVLFSGRTVRSALDALRDLGRPRAVQLAVLVDRGHRELP LRADYVGKNVPTARSEDVKVLLAEHDGRDAVIITSGQGAQT*
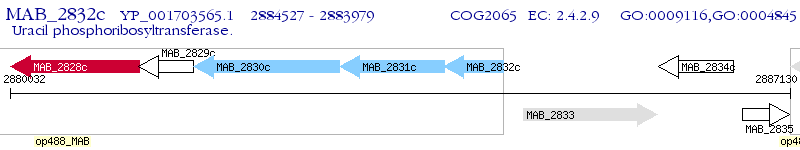
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2832c | - | - | 100% (182) | PyrR bifunctional protein (pyrimidine operon regulatory protein; Uracil phosphoribosyltransferase) |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1414 | pyrR | 2e-73 | 79.10% (177) | bifunctional pyrimidine regulatory protein PyrR uracil |
| M. gilvum PYR-GCK | Mflv_3748 | - | 1e-69 | 75.86% (174) | pyrimidine regulatory protein PyrR |
| M. tuberculosis H37Rv | Rv1379 | pyrR | 2e-73 | 79.10% (177) | bifunctional pyrimidine regulatory protein PyrR uracil |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_2194 | pyrR | 7e-73 | 78.86% (175) | pyrimidine operon regulatory protein PyrR |
| M. avium 104 | MAV_3394 | - | 3e-73 | 79.43% (175) | pyrimidine regulatory protein PyrR |
| M. smegmatis MC2 155 | MSMEG_3042 | - | 2e-72 | 77.71% (175) | pyrimidine regulatory protein PyrR |
| M. thermoresistible (build 8) | TH_1352 | pyrR | 3e-76 | 82.86% (175) | PROBABLE PYRIMIDINE OPERON REGULATORY PROTEIN PYRR |
| M. ulcerans Agy99 | MUL_1779 | pyrR | 5e-72 | 78.29% (175) | bifunctional pyrimidine regulatory protein PyrR uracil |
| M. vanbaalenii PYR-1 | Mvan_2659 | - | 1e-70 | 78.16% (174) | pyrimidine regulatory protein PyrR |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_3042|M.smegmatis_MC2_155 -----MGS-SSTDRELLSAADVGRTVSRIAHQIIEKTALDD--PAERTRV
TH_1352|M.thermoresistible__bu --VGSMADGSDAARELLSAADVGRTISRIAHQIIEKTALDG--P-DAPRV
Mb1414|M.bovis_AF2122/97 MGAAGDAAIGRESRELMSAADVGRTISRIAHQIIEKTALDDPVGPDAPRV
Rv1379|M.tuberculosis_H37Rv MGAAGDAAIGRESRELMSAADVGRTISRIAHQIIEKTALDDPVGPDAPRV
MAV_3394|M.avium_104 MGAAGNTGSSGDSRELMSAADVGRTVSRIAHQIIEKTALD---GPDGPRV
MMAR_2194|M.marinum_M MGAANDAVDG---RELMSAADVGRTISRIAHQIIEKTALD---SPDSPRV
MUL_1779|M.ulcerans_Agy99 MGAANDAVDG---RELMSAAVVGRTISRIAHQIIEKTALD---SPDSPRV
MAB_2832c|M.abscessus_ATCC_199 ---------------MLSAADVGRTISRIAHQIIEKTALSD--SGDAPRV
Mflv_3748|M.gilvum_PYR-GCK ---MGAPDQSGSDRELMSAADVSRTIARIAHQIIEKTALD---GPDGPRV
Mvan_2659|M.vanbaalenii_PYR-1 ---MGAPDQTGPDRELMSAADVSRTIARIAHQIIEKTALD---GPDGPRV
::*** *.**::************. : .**
MSMEG_3042|M.smegmatis_MC2_155 VLLGIPTRGVILATRLAAKIKEFAGEDVPHGALDITLYRDDLNFKPPRPL
TH_1352|M.thermoresistible__bu VLLGIPTRGVTLADRLATKIREFSGVQVPHGALDITLYRDDLNIKPPRPL
Mb1414|M.bovis_AF2122/97 VLLGIPTRGVTLANRLAGNITEYSGIHVGHGALDITLYRDDLMIKPPRPL
Rv1379|M.tuberculosis_H37Rv VLLGIPTRGVTLANRLAGNITEYSGIHVGHGALDITLYRDDLMIKPPRPL
MAV_3394|M.avium_104 VLLGIPTRGVTLADRLARNIGEYSGVEVGHGALDITLYRDDLMQKPPRPL
MMAR_2194|M.marinum_M VLLGIPTRGVILASRLAANIGEYSGIEVDHGALDITLYRDDLMSKPPRPL
MUL_1779|M.ulcerans_Agy99 VLLGIPTRGVILASRLAANIGEYSGIEVDHGALDITLYRDDLMSKPPRPL
MAB_2832c|M.abscessus_ATCC_199 VLVGIPTRGATLAKRLAAHITEFSGVEVPAGFLDITLYRDDLRNKPHRPL
Mflv_3748|M.gilvum_PYR-GCK ILLGIPTRGVTLASRLATNIAEFSGVQVAHGALDTTLYRDDLMTKPPRAL
Mvan_2659|M.vanbaalenii_PYR-1 ILLGIPTRGVTLATRLARNIAEFSGVEVAHGALDITLYRDDLMTKPPRAL
:*:******. ** *** :* *::* .* * ** ******* ** *.*
MSMEG_3042|M.smegmatis_MC2_155 EATSIPAGGVDDAIVILVDDVLYSGRSVRSALDALRDIGRPRIVQLAVLV
TH_1352|M.thermoresistible__bu EDTTIPEGGIDDALVILVDDVLFSGRSVRSALDALRDIGRPRAVQLAVLV
Mb1414|M.bovis_AF2122/97 ASTSIPAGGIDDALVILVDDVLYSGRSVRSALDALRDVGRPRAVQLAVLV
Rv1379|M.tuberculosis_H37Rv ASTSIPAGGIDDALVILVDDVLYSGRSVRSALDALRDVGRPRAVQLAVLV
MAV_3394|M.avium_104 EATSIPAGGIDDALVILVDDVLYSGRSVRSALDALRDVGRPRVVQLAVLV
MMAR_2194|M.marinum_M EETEIPAGGVDDALVILVDDVLYSGRSVRSALDALRDVGRPRSVQLAVLV
MUL_1779|M.ulcerans_Agy99 EETEIPAGGVDDALVILVDDVLYSGRSVRSALDALRDVGRPRSVQLAVLV
MAB_2832c|M.abscessus_ATCC_199 ERTSIPEGGVDGALVVLVDDVLFSGRTVRSALDALRDLGRPRAVQLAVLV
Mflv_3748|M.gilvum_PYR-GCK AETSIPAGGIDGALVILVDDVLYTGRSVRSALDALRDLGRPKAVQLAVLV
Mvan_2659|M.vanbaalenii_PYR-1 AATSIPEGGIDGALVVLVDDVLYSGRSVRSALDALRDLGRPKSVQLAVLV
* ** **:*.*:*:******::**:**********:***: *******
MSMEG_3042|M.smegmatis_MC2_155 DRGHRELPIRADYVGKNVPTSRSESVHVLLSEHDDRDGVVISK------
TH_1352|M.thermoresistible__bu DRGHRELPLRADYVGKNVPTSRSESVRVLLREHDDRDGVVISR------
Mb1414|M.bovis_AF2122/97 DRGHRELPLRADYVGKNVPTSRSESVHVRLREHDGRDGVVISR------
Rv1379|M.tuberculosis_H37Rv DRGHRELPLRADYVGKNVPTSRSESVHVRLREHDGRDGVVISR------
MAV_3394|M.avium_104 DRGHRELPLRAEYVGKNVPTSRSESVHVLLAEHDGADGVVISR------
MMAR_2194|M.marinum_M DRGHRELPVRADYVGKNVPTARSESVHVQFSEHDGHDGVVISR------
MUL_1779|M.ulcerans_Agy99 DRGHRELPVRADYVGKNVPTARSESVHVQFSEHDGHDGVVISR------
MAB_2832c|M.abscessus_ATCC_199 DRGHRELPLRADYVGKNVPTARSEDVKVLLAEHDGRDAVIITSGQGAQT
Mflv_3748|M.gilvum_PYR-GCK DRGHRELPIRADYVGKNVPTSRSENVKVRLTENDGVDIIAIAPYGGPPR
Mvan_2659|M.vanbaalenii_PYR-1 DRGHRELPIRADYVGKNVPTSRAENVKVRLTEDDGHDGVRIAPYGGPAR
********:**:********:*:*.*:* : *.*. * : *: