For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TKHRDSSLDDEQDSVEMLQAEIEEGSDEGESGVPVDPGEAEVLLEVKDLTLKFGGLQALDGVSFDIRRGE ILGLVGPNGAGKTTCFNAITGVYKPTSGSIVFNGKPLRRLRRHQITRRGIARTFQNIRLFGEMTALENVA VGTDARHKTSVLGALLRTPRHRREEHYAVDVGMGLLDFVGIADDADVKAKNLPYGSQRRLEIARALATEP KLLCLDEPAAGFTPSEKDSLIELIQRIRDYGYTVLLIEHDMKLVMGVTDRIVVLEFGRKIADGLPAEVRN DPAVIEAYLGVADEDFHGPGWLDSAPPDSESER*
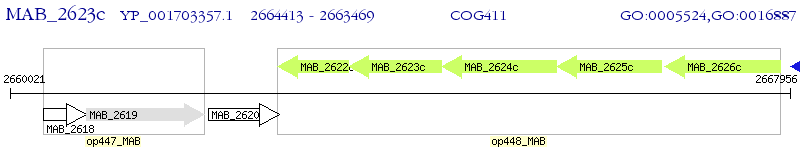
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2623c | - | - | 100% (314) | branched chain amino acid ABC transporter |
| M. abscessus ATCC 19977 | MAB_2622c | - | 2e-32 | 32.40% (250) | branched chain amino acid ABC transporter |
| M. abscessus ATCC 19977 | MAB_1655 | - | 1e-27 | 31.89% (254) | sulfate ABC transporter, ATP-binding protein SysA |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2419c | cysA1 | 2e-28 | 33.06% (248) | sulfate-transport ATP-binding protein ABC transporter CysA1 |
| M. gilvum PYR-GCK | Mflv_3571 | - | 1e-112 | 77.04% (257) | ABC transporter related |
| M. tuberculosis H37Rv | Rv2397c | cysA1 | 2e-28 | 33.06% (248) | sulfate-transport ATP-binding protein ABC transporter CysA1 |
| M. leprae Br4923 | MLBr_00669 | ftsE | 3e-24 | 32.26% (217) | putative cell division ATP-binding protein |
| M. marinum M | MMAR_3715 | cysA1 | 3e-29 | 33.07% (257) | sulfate-transport ATP-binding protein ABC transporter CysA1 |
| M. avium 104 | MAV_1785 | - | 4e-28 | 31.62% (253) | sulfate/thiosulfate import ATP-binding protein CysA |
| M. smegmatis MC2 155 | MSMEG_3250 | - | 1e-112 | 76.25% (261) | ABC transporter, ATP-binding protein |
| M. thermoresistible (build 8) | TH_3313 | - | 1e-113 | 78.60% (257) | PUTATIVE ABC transporter related |
| M. ulcerans Agy99 | MUL_3658 | cysA1 | 2e-29 | 33.07% (257) | sulfate-transport ATP-binding protein ABC transporter CysA1 |
| M. vanbaalenii PYR-1 | Mvan_2844 | - | 1e-113 | 78.21% (257) | ABC transporter-related protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2419c|M.bovis_AF2122/97 --------------------------------MT--------YAIVVADA
Rv2397c|M.tuberculosis_H37Rv --------------------------------MT--------YAIVVADA
MMAR_3715|M.marinum_M --------------------------MKKETSLTGPGT--GGHAIVVRDA
MUL_3658|M.ulcerans_Agy99 --------------------------------MTGPGT--GGHAIVVRDA
MAV_1785|M.avium_104 --------------------------------MTDAGTDRGDIAITVRDA
MLBr_00669|M.leprae_Br4923 -------------------------------------------MITLDHV
Mflv_3571|M.gilvum_PYR-GCK MTEP------------EAFEDAVAEIAGVHR---EIRADEGEVLLQTNEL
Mvan_2844|M.vanbaalenii_PYR-1 MTAPDNDAT--AETALENFEDSVADIAGVHR---EIQAEEGEVLLQTTDL
MSMEG_3250|M.smegmatis_MC2_155 MTEPP-----------QVAEEVIEDLAGVHR---EINAAEGEILLQTHDL
TH_3313|M.thermoresistible__bu --------------------MSIEELAGVHR---DIRAAEGEILLETVDL
MAB_2623c|M.abscessus_ATCC_199 MTKHRDSSLDDEQDSVEMLQAEIEEGSDEGESGVPVDPGEAEVLLEVKDL
: .
Mb2419c|M.bovis_AF2122/97 TKRYGDFV--ALDHVDFVVPTGSLTALLGPSGSGKSTLLRTIAGLDQPDT
Rv2397c|M.tuberculosis_H37Rv TKRYGDFV--ALDHVDFVVPTGSLTALLGPSGSGKSTLLRTIAGLDQPDT
MMAR_3715|M.marinum_M FKQYGDFV--ALDHVDFVVPAGSLTALLGPSGSGKSTLLRTIAGLDQPDS
MUL_3658|M.ulcerans_Agy99 FKQYGDFV--ALDHVDFVVPAGSLTALLGPSGSGKSTLLRTIAGLDQPDS
MAV_1785|M.avium_104 YKRYGDFV--ALDHVDFVVPTGSLTALLGPSGSGKSTLLRTIAGLDQPDT
MLBr_00669|M.leprae_Br4923 SKKYKSLARPALDNVNVKIDKGEFVFLIGPSGSGKSTFMRLLLGAETPTS
Mflv_3571|M.gilvum_PYR-GCK TVKFGGLT--ALDAVTFNIRRGEILGLIGPNGAGKTTCFNAITGVYRPTS
Mvan_2844|M.vanbaalenii_PYR-1 TVKFGGLT--ALDAVTFNIRRGEILGLIGPNGAGKTTCFNAITGVYRPTS
MSMEG_3250|M.smegmatis_MC2_155 TVKFGGLV--ALDSVNFGIRRGEILGLIGPNGAGKTTCFNAITGVYRPSS
TH_3313|M.thermoresistible__bu TVKFGGLV--ALDSVSFNIRRGEILGLIGPNGAGKTTCFNAITGVYRPAS
MAB_2623c|M.abscessus_ATCC_199 TLKFGGLQ--ALDGVSFDIRRGEILGLVGPNGAGKTTCFNAITGVYKPTS
:: .: *** * . : *.: *:**.*:**:* :. : * * :
Mb2419c|M.bovis_AF2122/97 GTITINGRDVTRVP-----PQRRGIGFVFQHYAAFKHLTVRDNVAFGLKI
Rv2397c|M.tuberculosis_H37Rv GTITINGRDVTRVP-----PQRRGIGFVFQHYAAFKHLTVRDNVAFGLKI
MMAR_3715|M.marinum_M GNITINGRDVTRVP-----PQRRGIGFVFQHYAAFKHLTVRDNVAYGLKI
MUL_3658|M.ulcerans_Agy99 GNITINGRDVTRVP-----PQRRGIGFVFQHYAAFKHLTVRDNVAYGLKI
MAV_1785|M.avium_104 GTVTIYGRDVTRVP-----PQRRGIGFVFQHYAAFKHLTVRDNVAYGLKV
MLBr_00669|M.leprae_Br4923 GDVRVSKFHVNKLPGRHIPRLRQVIGCVFQDFRLLQQKTVYENVAFALEV
Mflv_3571|M.gilvum_PYR-GCK GSVTFDGSTIGRIKR--HQITRLGIARTFQNIRLFGEMTALENVMVGTDA
Mvan_2844|M.vanbaalenii_PYR-1 GIVTFDGAPIGRIKR--HQITRLGIARTFQNIRLFGEMTALENVMVGTDA
MSMEG_3250|M.smegmatis_MC2_155 GSVTFDGAPLGRIKR--HQITRRGIARTFQNIRLFGEMTALENVMVGTDA
TH_3313|M.thermoresistible__bu GAVIFDGQKLGRIKR--HQITRLGIARTFQNIRLWGEMTALENVVVGTDA
MAB_2623c|M.abscessus_ATCC_199 GSIVFNGKPLRRLRR--HQITRRGIARTFQNIRLFGEMTALENVAVGTDA
* : . : :: * *. .**. . *. :** . .
Mb2419c|M.bovis_AF2122/97 RK---------------RPKAEIKAKVDNLLQVVGLSGFQSRYPNQLSGG
Rv2397c|M.tuberculosis_H37Rv RK---------------RPKAEIKAKVDNLLQVVGLSGFQSRYPNQLSGG
MMAR_3715|M.marinum_M RK---------------RPKAEIRDKVDNLLEVVGLSGFHGRYPNQLSGG
MUL_3658|M.ulcerans_Agy99 RK---------------RPKAEIRDKVDNLLEVVGLSGFHGRYPDQLSGG
MAV_1785|M.avium_104 RK---------------RPKAEIKAKVDNLLEVVGLSGFQGRYPNQLSGG
MLBr_00669|M.leprae_Br4923 IG---------------RRSDAINQVVPDVLETVGLSGKANRLPDELSGG
Mflv_3571|M.gilvum_PYR-GCK RHHTSVPGALIRSPRHRREERSAIERSAALLHFVGIAHRGEEKAKNLSYG
Mvan_2844|M.vanbaalenii_PYR-1 RHHTSVPGALIRSPRHRREERSAIERSAALLHFVGIAHRGEEKAKNLSYG
MSMEG_3250|M.smegmatis_MC2_155 RHRTSVPGALIRSPRHFREERSAVERSAALLHFVGIAHRGEEKAKNLSYG
TH_3313|M.thermoresistible__bu RHKTSVPGALFRSPRHRREERSAIERATALLHFVGIAHRAEEKAKNLPYG
MAB_2623c|M.abscessus_ATCC_199 RHKTSVLGALLRTPRHRREEHYAVDVGMGLLDFVGIADDADVKAKNLPYG
* . :*. **:: ..:*. *
Mb2419c|M.bovis_AF2122/97 QRQRMALARALAVDPEVLLLDEPFGALDAKVREELRAWLRRLHDEVHVTT
Rv2397c|M.tuberculosis_H37Rv QRQRMALARALAVDPEVLLLDEPFGALDAKVREELRAWLRRLHDEVHVTT
MMAR_3715|M.marinum_M QRQRMALARALAVDPEVLLLDEPFGALDAKVREDLRAWLRRLHDEVHVTT
MUL_3658|M.ulcerans_Agy99 QRQRMALARALAVDPEVLLLDEPFGALDAKVREDLRAWLRRLHDEVHVTT
MAV_1785|M.avium_104 QRQRMALARALAVDPQVLLLDEPFGALDAKVREDLRAWLRRLHDEVHVTT
MLBr_00669|M.leprae_Br4923 EQQRVAIARAFVNRPLVLLADEPTGNLDPDTSKDIMDLLERIN-RTGTTV
Mflv_3571|M.gilvum_PYR-GCK DQRRLEIARALATEPKLLCLDEPAAGFNPSEKSALIDLIRKIR-DDGYTV
Mvan_2844|M.vanbaalenii_PYR-1 DQRRLEIARALATEPKLLCLDEPAAGFNPSEKSALIELIRKIR-DDGYTV
MSMEG_3250|M.smegmatis_MC2_155 DQRRLEIARALATEPKLLCLDEPAAGFNPSEKAALIDLIRKIR-DDGYTV
TH_3313|M.thermoresistible__bu DQRRLEIARALATEPKLLCLDEPAAGFNPSEKAALIDLIRKIR-DDGYTV
MAB_2623c|M.abscessus_ATCC_199 SQRRLEIARALATEPKLLCLDEPAAGFTPSEKDSLIELIQRIR-DYGYTV
.::*: :***:. * :* *** . : .. : :.::. *.
Mb2419c|M.bovis_AF2122/97 VLVTHDQAEALDVADRIAVLHKGRIEQVGSPTDVYDAPANAFVMSFLGAV
Rv2397c|M.tuberculosis_H37Rv VLVTHDQAEALDVADRIAVLHKGRIEQVGSPTDVYDAPANAFVMSFLGAV
MMAR_3715|M.marinum_M VLVTHDQAEALDVADRIAVLNSGRIEQVGSPTEVYDAPANAFVMSFLGAV
MUL_3658|M.ulcerans_Agy99 VLVTHDQAEALDVADRIAVLNSGRIEQVGSPTEVYDAPTNAFVMSFLGAV
MAV_1785|M.avium_104 VLVTHDQAEALDVADRIAVLNQGRIEQIGSPTEVYDAPTNAFVMSFLGAV
MLBr_00669|M.leprae_Br4923 LMATHDHHIVDSMRQRVVELSLG---------------------------
Mflv_3571|M.gilvum_PYR-GCK LLIEHDMRLVMGVTDRIVVLEFGRKIADGLPSEIREDP------------
Mvan_2844|M.vanbaalenii_PYR-1 LLIEHDMRLVMGVTDRIVVLEFGRKIADGLPAEIREDP------------
MSMEG_3250|M.smegmatis_MC2_155 LLIEHDMRLVMGVTDRIVVLEFGRKIADGLPSQIREDP------------
TH_3313|M.thermoresistible__bu LLIEHDMKLVMGVTDRIVVLEFGRKIADGPPAAIREDP------------
MAB_2623c|M.abscessus_ATCC_199 LLIEHDMKLVMGVTDRIVVLEFGRKIADGLPAEVRNDP------------
:: ** . .: :*:. * *
Mb2419c|M.bovis_AF2122/97 STLNGSLVRPHDIRVGRTPNMAVAAADGTAGSTGVLRAVVDRVVVLGFEV
Rv2397c|M.tuberculosis_H37Rv STLNGSLVRPHDIRVGRTPNMAVAAADGTAGSTGVLRAVVDRVVVLGFEV
MMAR_3715|M.marinum_M SALNGALVRPHDIRVGRTPEMAVAAADGTGESTGVTRAVVDRIVVLGFEV
MUL_3658|M.ulcerans_Agy99 SALNGALVRPHDIRVGRTPEMAVAAADGTGESTGVTRAVVDRIVVLGFEV
MAV_1785|M.avium_104 STLNGTLVRPHDIRVGRTPEMAVAAEDGTAESTGVARAIVDRVVKLGFEV
MLBr_00669|M.leprae_Br4923 -------------RLVRDEWCGIYGMDR----------------------
Mflv_3571|M.gilvum_PYR-GCK --------------KVIAAYLGVPDDELA---------------------
Mvan_2844|M.vanbaalenii_PYR-1 --------------KVIAAYLGVPDDELT---------------------
MSMEG_3250|M.smegmatis_MC2_155 --------------KVIAAYLGVPDDEIR---------------------
TH_3313|M.thermoresistible__bu --------------AVIAAYLGVPDDEIG---------------------
MAB_2623c|M.abscessus_ATCC_199 --------------AVIEAYLGVADEDFHGPGWLDSAPPDSESER-----
.: :
Mb2419c|M.bovis_AF2122/97 RVELTSAATGGAFTAQITRGDAEALALREGDTVYVRATRVPPIAG---GV
Rv2397c|M.tuberculosis_H37Rv RVELTSAATGGAFTAQITRGDAEALALREGDTVYVRATRVPPIAG---GV
MMAR_3715|M.marinum_M RVELTSAATGGAFTAQITRGDAEALALQEGDTVYVRATRVPPIAG---AA
MUL_3658|M.ulcerans_Agy99 RVELTSAATGGAFTAQITRGDAEALALQEGDTVYVRATRVPPIAG---AA
MAV_1785|M.avium_104 RVELTSAATGGPFTAQITRGDAEALALREGDTVYVRATRVPPITAGATTV
MLBr_00669|M.leprae_Br4923 --------------------------------------------------
Mflv_3571|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2844|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_3250|M.smegmatis_MC2_155 --------------------------------------------------
TH_3313|M.thermoresistible__bu --------------------------------------------------
MAB_2623c|M.abscessus_ATCC_199 --------------------------------------------------
Mb2419c|M.bovis_AF2122/97 SGVDDAGVERVKVTST------
Rv2397c|M.tuberculosis_H37Rv SGVDDAGVERVKVTST------
MMAR_3715|M.marinum_M PAVPDDGAGSTPSAAQIGVGGQ
MUL_3658|M.ulcerans_Agy99 PAVPDDGAGSTPRAAQIGVGGQ
MAV_1785|M.avium_104 PAVSRDGADEATLTSA------
MLBr_00669|M.leprae_Br4923 ----------------------
Mflv_3571|M.gilvum_PYR-GCK ----------------------
Mvan_2844|M.vanbaalenii_PYR-1 ----------------------
MSMEG_3250|M.smegmatis_MC2_155 ----------------------
TH_3313|M.thermoresistible__bu ----------------------
MAB_2623c|M.abscessus_ATCC_199 ----------------------