For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MVRELGAGGWRDLLDGRQLGIVTVLAGGVALYATNVYLTTSLLPSAVVEIGGERFYAWVTTVYLIASVSA ATVVSPLLGWAGPRGAYLLAFGAFAVGTLVCAVAPGMHVLLAGRAVQGAAGGLLAGLAYAVINLALPQGL WTRASALTSAMWGVGTFVGPAAGGLFAQWGLWRWAFGMLAVITAGIAAMVPYVLTRAQKPNAARAGNQVP VWSLLVLTAAASLVAAAGLQSTLPFMATLLAAAVVLVILFVALDRRSRASVLPKAVFARNPLKWIYLTIA VLAVASMAETYIPFFGQRLGHLSPVAAGFLGATLAFGWTVGEMISASAQVRTVSRVVSVAPAVVAAGLFL AGVLQRDGATVLEIGCWALGFVVTGAGIGMAWPHLAAAAMSVSDDGSESGTGAAAISTVQLIAGALGAGL AGVFVNLGGSARHGAQLLFVGFAVMVLCGVVVSVRVFRTGSASEMTDRAGQQAVRIPS
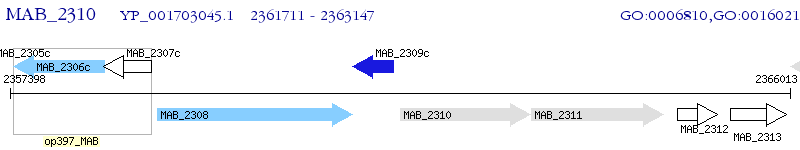
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2310 | - | - | 100% (478) | drug efflux membrane protein |
| M. abscessus ATCC 19977 | MAB_0970c | - | 3e-29 | 25.78% (419) | drug resistance transporter |
| M. abscessus ATCC 19977 | MAB_0945 | - | 4e-20 | 25.93% (378) | EmrB/QacA family drug resistance transporter |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1660 | - | 1e-133 | 54.80% (458) | drug efflux membrane protein |
| M. gilvum PYR-GCK | Mflv_3321 | - | 2e-30 | 27.25% (411) | EmrB/QacA family drug resistance transporter |
| M. tuberculosis H37Rv | Rv1634 | - | 1e-132 | 54.87% (452) | drug efflux membrane protein |
| M. leprae Br4923 | MLBr_00556 | - | 1e-15 | 23.83% (428) | putative integral membrane drug transport protein |
| M. marinum M | MMAR_2438 | - | 1e-129 | 53.20% (453) | drug efflux membrane protein |
| M. avium 104 | MAV_3140 | - | 1e-137 | 54.84% (465) | drug efflux membrane protein |
| M. smegmatis MC2 155 | MSMEG_3815 | - | 1e-139 | 57.27% (454) | drug efflux membrane protein |
| M. thermoresistible (build 8) | TH_2729 | - | 8e-27 | 25.70% (463) | PUTATIVE putative drug resistance transporter, EmrB/QacA |
| M. ulcerans Agy99 | MUL_1617 | - | 1e-128 | 52.98% (453) | drug efflux membrane protein |
| M. vanbaalenii PYR-1 | Mvan_3349 | - | 1e-140 | 57.14% (462) | major facilitator superfamily transporter |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_3815|M.smegmatis_MC2_155 MTETAA---AEAAGTWRQLLGRRYLGASTVLAGGVLLYATNEFITISLLP
Mvan_3349|M.vanbaalenii_PYR-1 MTHTEAGSAAVATGSWRELLGPKYLGASTVLAGGVALYATNEFLTISLMP
Mb1660|M.bovis_AF2122/97 MTETAS-----ETGSWRELLSR-YLGTSIVLAGGVALYATNEFLTISLLP
Rv1634|M.tuberculosis_H37Rv MTETAS-----ETGSWRELLSR-YLGTSIVLAGGVALYATNEFLTISLLP
MMAR_2438|M.marinum_M MTETAR-----ETGSWRELLSG-HLGTATLLAGGVAMYATNEFLTVSLLP
MUL_1617|M.ulcerans_Agy99 MTETAR-----ETGSWRELLRG-HLGTATLLAGGVAMYATNEFLTVSLLP
MAV_3140|M.avium_104 MTDAAT-----ITGGWRELLGARYLRTSILLAGGVALYATNEFLTTSLLP
MAB_2310|M.abscessus_ATCC_1997 MVRELG------AGGWRDLLDGRQLGIVTVLAGGVALYATNVYLTTSLLP
Mflv_3321|M.gilvum_PYR-GCK ----------------------------------MLLAALDQTIVATALP
TH_2729|M.thermoresistible__bu --------------------------VLVALILTMALVAMDTTILATAVP
MLBr_00556|M.leprae_Br4923 MSTRAG-----------------RRVAISAGSLAVLLGALDTYVVVTIMR
: : * : : : :
MSMEG_3815|M.smegmatis_MC2_155 SAVADIGG----QRFYAWVTTVYLVASVVAATTVHSVLMRLGPRVAYLVA
Mvan_3349|M.vanbaalenii_PYR-1 SAVADIGG----HRFYAWVTTVYLVGSVMAATTVHSVLTRLGPRWAYLLG
Mb1660|M.bovis_AF2122/97 STIADIGG----SRLYAWVTTLYLVGSVVAATTVNTMLLRVGARSSYLMG
Rv1634|M.tuberculosis_H37Rv STIADIGG----SRLYAWVTTLYLVGSVVAATTVNTMLLRVGARSSYLMG
MMAR_2438|M.marinum_M STIAEIGG----SRLYAWVTTLYLVGSVVSATTVNAMLLKVGARMSYLMG
MUL_1617|M.ulcerans_Agy99 STIAEIGG----SRLYAWVTTLYLVGSVVSATTVNAMLLKVGARMSYLMG
MAV_3140|M.avium_104 NTIAEIGG----SRLYAWVTTLYLVGSVVAATMVNPMLLRVGARASYLLG
MAB_2310|M.abscessus_ATCC_1997 SAVVEIGG----ERFYAWVTTVYLIASVSAATVVSPLLGWAGPRGAYLLA
Mflv_3321|M.gilvum_PYR-GCK TVVADLGG----AGHQSWVVTSYLLASTIATAIVGKLGDLFGRKMIFGVA
TH_2729|M.thermoresistible__bu QVVGDLGG----FDQVGWVFSGYLLAQTVTIPIYGKLADLYGRRPILIFG
MLBr_00556|M.leprae_Br4923 DIMHDVGIPVNQMQRITWIVTMYLLGYIAAMPLLSRASDRFGRKLLLQVS
: ::* *: : **:. : . * : ..
MSMEG_3815|M.smegmatis_MC2_155 LTVFGAGSLGCALA---PSMQLLLAGRLVQGFAGGLLAGLGYAVINTALP
Mvan_3349|M.vanbaalenii_PYR-1 LTVFGLGSLVCAVA---PSMEILLAGRTVQGAAGGLLAGLGYAVINTVLP
Mb1660|M.bovis_AF2122/97 LAVFGLASLVCAAA---PSMQILVAGRTLQGIAGGLLAGLGYALINSTLP
Rv1634|M.tuberculosis_H37Rv LAVFGLASLVCAAA---PSMQILVAGRTLQGIAGGLLAGLGYALINSTLP
MMAR_2438|M.marinum_M LTVFGFASLVCAAA---QNMEMLVAGRTLQGVAGGLLAGLGYALINAALP
MUL_1617|M.ulcerans_Agy99 LTVFGFASLVCAAA---QNMEMLVAGRTLQGVAGGLLAGLGYALINAALP
MAV_3140|M.avium_104 LAVFGVASLVCGAA---PDMHVLIAGRAMQGVAGGLLAGLGYAVINSALP
MAB_2310|M.abscessus_ATCC_1997 FGAFAVGTLVCAVA---PGMHVLLAGRAVQGAAGGLLAGLAYAVINLALP
Mflv_3321|M.gilvum_PYR-GCK ILFFLAGSVLCGIA---STMEMLVASRALQGIGGGALMVTATALIGEVIP
TH_2729|M.thermoresistible__bu VVAFLIGSALSAAS---WNMATLAIFRAIQGLGAGAISATVQTVAGDLYT
MLBr_00556|M.leprae_Br4923 LAGFAIGSVMTALAGQFGDFHMLIAGRTIQGVASGALLPITLALGADLWA
. * .: . : : * * :** ..* : :: .
MSMEG_3815|M.smegmatis_MC2_155 NTLWTKASALVSAMWGVGTVVGPASGGLFAQYG-SWRWAFGVLVILTAAM
Mvan_3349|M.vanbaalenii_PYR-1 SVLWKKASALVSAMWGVGTLVGPAAGGLFAQYS-SWRWAFGILVVMTTAM
Mb1660|M.bovis_AF2122/97 KSLWTRGSALVSAMWGVATLIGPATGGLFAQLG-LWRWAFGVMTLLTALM
Rv1634|M.tuberculosis_H37Rv KSLWTRGSALVSAMWGVATLIGPATGGLFAQLG-LWRWAFGVMTLLTALM
MMAR_2438|M.marinum_M HNLWTRGSALVSAMWGVATLIGPATGGLFAQFG-LWRWAFGVMATMAGLM
MUL_1617|M.ulcerans_Agy99 HNLWTCGSALVSAMWGVATLIGPATGGLFAQFG-LWRWAFGVMATMAGLM
MAV_3140|M.avium_104 RWLWTRGSALVSAMWGVATVVGPATGGLFAQLG-IWRWAFVVMAVLTALM
MAB_2310|M.abscessus_ATCC_1997 QGLWTRASALTSAMWGVGTFVGPAAGGLFAQWG-LWRWAFGMLAVITAGI
Mflv_3321|M.gilvum_PYR-GCK LRDRGRYQGALGAVFGVTTVIGPLLGGYFTDHL-TWRWAFWINVPIGIVV
TH_2729|M.thermoresistible__bu VAERGRIQGYLASVWGISAVIAPALGGFFAQYL-TWRWIFLVNIPIGVFA
MLBr_00556|M.leprae_Br4923 QRNRAGVLGGIGAAQELGSVLGPLYGIFIVWLFSDWRYVFWINIPLTAIA
. .: : :.:.* * :. **: * : :
MSMEG_3815|M.smegmatis_MC2_155 AVLVPLALPGRG-----SDTLVPPG-IPVWSLVLLGSAAMAVSVAGIPHD
Mvan_3349|M.vanbaalenii_PYR-1 SALVPMALPARARAA-HLGSGAPPGRIPFWSLLLLGAAALLVSVAGIPHD
Mb1660|M.bovis_AF2122/97 AMLVPVALGAGRVGPGGETPVGSTHKVPVWSLLLMGAAALAISVAALPNY
Rv1634|M.tuberculosis_H37Rv AMLVPVALGAGGVGPGGETPVGSTHKVPVWSLLLMGAAALAISVAALPNY
MMAR_2438|M.marinum_M TLLVLVVLSNG-VAVGGEKPVAPAHKVPVWSLLLMGAAALAVSVAEIPQY
MUL_1617|M.ulcerans_Agy99 TLLVLVVLSNG-VAVGGEKPAAPAHKVPVWSLLLMGAAALAVSVAEIPQY
MAV_3140|M.avium_104 ALLVPVALARV-----DPAPAIPRMKVPVWSLLIIGVASLAVSVAQIPHN
MAB_2310|M.abscessus_ATCC_1997 AAMVPYVLTRAQ----KPNAARAGNQVPVWSLLVLTAAASLVAAAGLQST
Mflv_3321|M.gilvum_PYR-GCK LIVALATIPALSRRGKPVIDYAGIVLIGLGASALT--LATSWGGSTYAWS
TH_2729|M.thermoresistible__bu LYLIARDLHEDVVRRPHRIDYLGAGLVLVSAGLFI--LGLLSGGVHWSWT
MLBr_00556|M.leprae_Br4923 MLMIQVSLSAHDRGDELEKVDVVGGVLLAIALGLVVIGLYNPQPDSKQVL
: : : : .
MSMEG_3815|M.smegmatis_MC2_155 PRGTAALLLVGVALVVAFIVVDRRAGASVLPPSTFG-PGPLKWIYMSLGV
Mvan_3349|M.vanbaalenii_PYR-1 VRATAGLVVAGFVLVAVFVVVDRRVAASVLPPSAFG-RGPLKWIYLTLGG
Mb1660|M.bovis_AF2122/97 LVQTAGLLAAAALLVAVFVVVDWRIHAAVLPPSVFG-SGPLKWIYLTMSV
Rv1634|M.tuberculosis_H37Rv LVQTAGLLAAAALLVAVFVVVDWRIHAAVLPPSVFG-SGPLKWIYLTMSV
MMAR_2438|M.marinum_M LVATAGLLVAGALLVGIFVIVDWRMHVAVLPHSVFG-SGPLKWIYLTMSV
MUL_1617|M.ulcerans_Agy99 LVATAGLLVAGALLVGIFVIVDWRMHVAVLPHSVFG-SGPLKWIYLTMSV
MAV_3140|M.avium_104 TAATFGLLAAGIMLVGLFVIVDWRMHAAILPPSVFS-PGPLKWIYLTMGV
MAB_2310|M.abscessus_ATCC_1997 LPFMATLLAAAVVLVILFVALDRRSRASVLPKAVFA-RNPLKWIYLTIAV
Mflv_3321|M.gilvum_PYR-GCK SPMIIGLFVGSAVALAVFVRVELRATEPVLPIRLFASPVFTVCCVLSFIV
TH_2729|M.thermoresistible__bu SPTSVVVFTGAALAGVALLVVESRAAEPVLPLSMLTRRLTAPSFAATATA
MLBr_00556|M.leprae_Br4923 PSYGVPVLVGGIVATVAFAVWERCARTRLIDPAGVHIRPFLSALGASVAA
:. . : : :: . :
MSMEG_3815|M.smegmatis_MC2_155 LMAATMVDMYVPLFGQRLAHMTPVVAGFFGAVLSVGWTFGEIASASLQNR
Mvan_3349|M.vanbaalenii_PYR-1 LMAATMVDMYVPLFGQRLAHLTPVAAGFLGAGLAIGWTVGEIGSASLTRH
Mb1660|M.bovis_AF2122/97 QMIAAMVDTYVPLFGQRLGHLTPVAAGFLGAALAVGWTVGEVASASLNSA
Rv1634|M.tuberculosis_H37Rv QMIAAMVDTYVPLFGQRLGHLTPVAAGFLGAALAVGWTVGEVASASLNSA
MMAR_2438|M.marinum_M QMVAAMVTTYVPLFGQGLGHLTPVAAGFLGAALAFGWTAGEIVSASLTNP
MUL_1617|M.ulcerans_Agy99 QMVAAMVTTYVPLFGQGLGHLTPVAAGLLGAALAFGWTAGEIVSASLTNS
MAV_3140|M.avium_104 LMAAAMVNTYVPLFGQRLAHLTPIAAGFLGAALALGWTVSEIVSASPENP
MAB_2310|M.abscessus_ATCC_1997 LAVASMAETYIPFFGQRLGHLSPVAAGFLGATLAFGWTVGEMISAS-AQV
Mflv_3321|M.gilvum_PYR-GCK GFAMLGALTFLPTFMQFVDGVSATESGLRTLPMVGGLLLTSMGSGVLVSR
TH_2729|M.thermoresistible__bu GMIVIGLSVYLPNWGQTVLGLSPVAAGFVLAIMSITWPISSGFSARLYLR
MLBr_00556|M.leprae_Br4923 GAALMVTLVNVELFGQGVLGMDQTQAAGLLVWFLIALPIGAVLGGWGATK
: : * : : :. : ..
MSMEG_3815|M.smegmatis_MC2_155 KVIRRMVAVAPLVMALGLAAG-------AVLVRNGMPVWLVVLWALALLV
Mvan_3349|M.vanbaalenii_PYR-1 RVVVRTVAVAPAVMATGLTIG-------ALTQHRDAGPMLVALWAVGLII
Mb1660|M.bovis_AF2122/97 RVIGHVVAAAPLVMASGLALG-------AVTQRADAPVGIIALWALALLI
Rv1634|M.tuberculosis_H37Rv RVIGHVVAAAPLVMASGLALG-------AVTQRADAPVGIIALWALALLI
MMAR_2438|M.marinum_M RLIGYIIAAAPLVMASGLALG-------AVTQRADASLPTVALWALALLI
MUL_1617|M.ulcerans_Agy99 RLIGYIIAVAPLVMASGLALG-------AVTQRADASLPTVALWALALLI
MAV_3140|M.avium_104 RTVGRVVMVAPLVAASGLALG-------AVARHGDGSAWTAALWAVALLV
MAB_2310|M.abscessus_ATCC_1997 RTVSRVVSVAPAVVAAGLFLA-------GVLQRDGATVLEIGCWALGFVV
Mflv_3321|M.gilvum_PYR-GCK TGRYKIFPVLGTAVMTVGFVL-------LSQMDATTPVWQQSAY---LFV
TH_2729|M.thermoresistible__bu IG-FRDTALVGASCAVAAGTG-------LVLLGPAPPVWQPVVY---TAL
MLBr_00556|M.leprae_Br4923 AGDRTMTFVGLLITAGGYWLISHWPVDLLNYRRSIFGLFSVPTMYADLLV
:
MSMEG_3815|M.smegmatis_MC2_155 TGAGIGIAWPHLSAWAMSSVDDPAEGPAAAAAINTVQLICGAFGAGLAG-
Mvan_3349|M.vanbaalenii_PYR-1 TGAGVGIAWPHLSAWAMSKVDDPAEGPAAAAAINTVQVISAAFGAALAG-
Mb1660|M.bovis_AF2122/97 IGTGIGIAWPHLTVRAMDSVADPAESSAAAAAINVVQLISGAFGAGLAG-
Rv1634|M.tuberculosis_H37Rv IGTGIGIAWPHLTVRAMDSVADPAESSAAAAAINVVQLISGAFGAGLAG-
MMAR_2438|M.marinum_M TGAGIGIAWPHLTVRAMDCVDDPAESSSAAAAINIVQLVSAAFGAGLAG-
MUL_1617|M.ulcerans_Agy99 TGAGIGIAWPHLTVRAMDCVDDPAESSSAAAAINIVQLVSAAFGAGLAG-
MAV_3140|M.avium_104 AGTGIGMAWPHLSARAMASVNDPAEGGAASAAINTVQLTSAAIGAGLAG-
MAB_2310|M.abscessus_ATCC_1997 TGAGIGMAWPHLAAAAMSVSDDGSESGTGAAAISTVQLIAGALGAGLAG-
Mflv_3321|M.gilvum_PYR-GCK LGTGIGMCMQVLVLIVQSTAN-FSDLGVATSGVTFFRTIGSSFGAAIFGS
TH_2729|M.thermoresistible__bu MGAGMGLLFSPLIVGLQNTVG-WDQRGTVTGGLMFSRFLGQSIGAAGFG-
MLBr_00556|M.leprae_Br4923 AGLGLGLVIGPLSSATLRVVP-TAQHGIASAAVVVARMTGMLIGVAALTA
* *:*: * : :..: : :*..
MSMEG_3815|M.smegmatis_MC2_155 --------------------------------VVVNLVNVGNPSSATWLF
Mvan_3349|M.vanbaalenii_PYR-1 --------------------------------VIVNLSHTGDVAAARWLF
Mb1660|M.bovis_AF2122/97 --------------------------------VVVNTAKGGEVAAARGLY
Rv1634|M.tuberculosis_H37Rv --------------------------------VVVNTAKGGEVAAARGLY
MMAR_2438|M.marinum_M --------------------------------VVVNTATGGEVAAARWLY
MUL_1617|M.ulcerans_Agy99 --------------------------------VVVNTATGGEVAAARWLY
MAV_3140|M.avium_104 --------------------------------VVVNTATGGDEMAAHLLF
MAB_2310|M.abscessus_ATCC_1997 --------------------------------VFVNLG-GSARHGAQLLF
Mflv_3321|M.gilvum_PYR-GCK LFANFLDSRIGGALAASGAPPEAAQSPQVLHRLPDAVAAPIVEAYSDSLG
TH_2729|M.thermoresistible__bu -----------------------AVANAVLRRYEPDTSAAAMGAASHAVF
MLBr_00556|M.leprae_Br4923 WGLYRFNQILAG-------LSTAVPPDATLIERAAAVADQAKRAYTMMYS
:
MSMEG_3815|M.smegmatis_MC2_155 AAFAVLAVAGTLASIRAVR-------------------------------
Mvan_3349|M.vanbaalenii_PYR-1 ASFAVLAVLATVASTRSGRLRSR---------------------------
Mb1660|M.bovis_AF2122/97 MAFTVLAAAGVIASYQATHRDRRLPR------------------------
Rv1634|M.tuberculosis_H37Rv MAFTVLAAAGVIASYQATHRDRRLPR------------------------
MMAR_2438|M.marinum_M TVFTVLAAVGFIASYQATHHDRRSPG------------------------
MUL_1617|M.ulcerans_Agy99 TVFTVLAAVGFIASYQATHHDRRSPG------------------------
MAV_3140|M.avium_104 TVFTALSAAGVAVSYAATRATRQAQPVGNVG-------------------
MAB_2310|M.abscessus_ATCC_1997 VGFAVMVLCGVVVSVRVFRTGSASEMTDRAGQQAVRIPS-----------
Mflv_3321|M.gilvum_PYR-GCK LVFLCAAPVAVIGFVVALTLKQVPLREIEATAALDMGEGFGMPSDDSSER
TH_2729|M.thermoresistible__bu VGLLVAAVATVVLLLTVP--RRFPTHHVAVVD------------------
MLBr_00556|M.leprae_Br4923 DIFMITAIVCVIGALLGLLISSRKEHASEPKVPE----------------
:
MSMEG_3815|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_3349|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb1660|M.bovis_AF2122/97 --------------------------------------------------
Rv1634|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2438|M.marinum_M --------------------------------------------------
MUL_1617|M.ulcerans_Agy99 --------------------------------------------------
MAV_3140|M.avium_104 --------------------------------------------------
MAB_2310|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_3321|M.gilvum_PYR-GCK MMETAVGRLVRHSPEIRLRSIAGTHGCDPDVALLWALIQIYRQSQVFGSA
TH_2729|M.thermoresistible__bu --------------------------------------------------
MLBr_00556|M.leprae_Br4923 --------------------------------------------------
MSMEG_3815|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_3349|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb1660|M.bovis_AF2122/97 --------------------------------------------------
Rv1634|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2438|M.marinum_M --------------------------------------------------
MUL_1617|M.ulcerans_Agy99 --------------------------------------------------
MAV_3140|M.avium_104 --------------------------------------------------
MAB_2310|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_3321|M.gilvum_PYR-GCK RLSEIAERLRVPPEVLAPTFDRLVATGRALRTDDQLWLTQSGAQQVSAAT
TH_2729|M.thermoresistible__bu --------------------------------------------------
MLBr_00556|M.leprae_Br4923 --------------------------------------------------
MSMEG_3815|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_3349|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb1660|M.bovis_AF2122/97 --------------------------------------------------
Rv1634|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2438|M.marinum_M --------------------------------------------------
MUL_1617|M.ulcerans_Agy99 --------------------------------------------------
MAV_3140|M.avium_104 --------------------------------------------------
MAB_2310|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_3321|M.gilvum_PYR-GCK AALVSRIVEKLEKSPTFEGRPDWDEVEKALERIAQRLLVQREWEDDRTEL
TH_2729|M.thermoresistible__bu --------------------------------------------------
MLBr_00556|M.leprae_Br4923 --------------------------------------------------
MSMEG_3815|M.smegmatis_MC2_155 -----
Mvan_3349|M.vanbaalenii_PYR-1 -----
Mb1660|M.bovis_AF2122/97 -----
Rv1634|M.tuberculosis_H37Rv -----
MMAR_2438|M.marinum_M -----
MUL_1617|M.ulcerans_Agy99 -----
MAV_3140|M.avium_104 -----
MAB_2310|M.abscessus_ATCC_1997 -----
Mflv_3321|M.gilvum_PYR-GCK AAAGK
TH_2729|M.thermoresistible__bu -----
MLBr_00556|M.leprae_Br4923 -----