For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AGRPPLRIGQHGKISRQYVGGGVWVARCRYRDSDGVTRIVERRGPVDDFDKHGKLAEDALIEALSDRRLP ADTDTIGYDTLVMALVDQHIGRLSDDGRSPATVATYKFASDKLRNFLGGVRVSEAHAARMDAALRSMRTA HGTTMARQAKTILQGGLQLAVMANVIPANPVRDVQSLRSKTRPKGATALSADQLRTLLAEIRASEYCAGN DLADPLIILMATGLRRSELLALRWSDFDANAGTLTVTGKLVRATGAGLQRIDEAKTAAGMRTVALPSFAV TALRHRQGRDYVGQQLMIFPSTAGTWRDPNNFGKQWRAVRDDLGVPGVTTHSFRKTMATLIDDNGLSARV GADQLGHAKVSMTQDRYMARGKVHHEVADLLDATISDE*
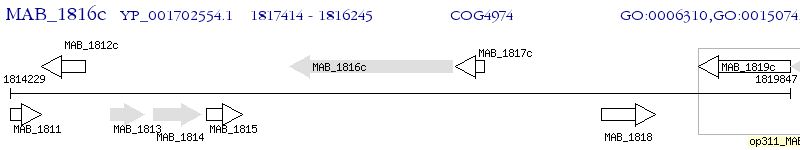
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1816c | - | - | 100% (389) | Phage integrase |
| M. abscessus ATCC 19977 | MAB_0221c | - | 2e-28 | 31.42% (331) | putative phage integrase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2786 | - | 5e-14 | 29.49% (217) | phage integrase family protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_01365 | xerD | 5e-06 | 27.41% (270) | site-specific tyrosine recombinase XerD |
| M. marinum M | MMAR_3607 | - | 1e-147 | 65.03% (386) | phage integrase |
| M. avium 104 | MAV_0253 | - | 1e-13 | 26.77% (325) | site-specific recombinase, phage integrase family protein |
| M. smegmatis MC2 155 | MSMEG_5754 | - | 1e-12 | 30.56% (216) | gp41 protein |
| M. thermoresistible (build 8) | TH_1542 | xerD | 5e-05 | 27.31% (260) | tyrosine recombinase XerD |
| M. ulcerans Agy99 | MUL_3581 | - | 1e-27 | 61.54% (91) | phage-related integrase |
| M. vanbaalenii PYR-1 | Mvan_3747 | - | 3e-14 | 29.17% (216) | phage integrase family protein |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_1816c|M.abscessus_ATCC_199 ------------------------------------MAGRPPLRIGQHGK
MMAR_3607|M.marinum_M -------MQSGDSQRWNNPWPGKSWPGMSSLAEVGTMAGRPQLRIGTHGQ
MUL_3581|M.ulcerans_Agy99 --------------------------------------------------
Mflv_2786|M.gilvum_PYR-GCK MQATTVQTPAGAWMTTTDKPPPKRTRRTEPITRRNAKNGAVSYTFQVDVG
Mvan_3747|M.vanbaalenii_PYR-1 MQATTVQTPAGAWMTTTDKPPPKRTRRTEPIMRRNAKNGAVSYTFQVDVG
MAV_0253|M.avium_104 ------------------------MAKIPDYVKALDLDSGRRYEVRPEV-
MSMEG_5754|M.smegmatis_MC2_155 --------------------MLQRMARIPDYVKVIDLDSGRRYEVRPEV-
MLBr_01365|M.leprae_Br4923 --------------------------------------------------
TH_1542|M.thermoresistible__bu --------------------------------------------------
MAB_1816c|M.abscessus_ATCC_199 ISRQYVGGGVWVARCRYRDSDGVTRIVERRGPVDDFDKHGKLAEDALIEA
MMAR_3607|M.marinum_M IKRIYTGGGVWLARCRYRDTDGVTRIVQRLGPADEYDQHGKLAADALIAA
MUL_3581|M.ulcerans_Agy99 --------------------------------------------------
Mflv_2786|M.gilvum_PYR-GCK THADGTRDRQRFTFSTFAEARREYRKISTEVAAGTFVKRDLTTVAEYLAR
Mvan_3747|M.vanbaalenii_PYR-1 THADGTRDRQRFTFSTFAEARREYRKISTEVAAGTFVKRDLTTVAEYLGR
MAV_0253|M.avium_104 TGPDGKRKQQRKRFKTLKEATDFYTRIAGDRSRGVHVAPSELTVEAAVDA
MSMEG_5754|M.smegmatis_MC2_155 TGADGKRRQKRERFKTLKEAVDHYTAIAGDRGRGVHVAPSELTVEDAVDS
MLBr_01365|M.leprae_Br4923 --------------------MTT--------------AALQTQLQGYLD-
TH_1542|M.thermoresistible__bu --------------------VTTSEQVPRSDEPPHPHRVLDDQIQGYLD-
MAB_1816c|M.abscessus_ATCC_199 LSDRRLPADTDTIGYDTLVMALVDQHIGRLSDDGRSPATVATYKFASDKL
MMAR_3607|M.marinum_M LAQRRHSANHGEIGPDTKVTDLVEQHLSRLAEDGRSPVTMSTYRFTTKNL
MUL_3581|M.ulcerans_Agy99 --------------------------------------------------
Mflv_2786|M.gilvum_PYR-GCK WLDGRRDVRVNTLAGYRHSLKPVIDNLGGLGLQQLRTS---DIDALVTLR
Mvan_3747|M.vanbaalenii_PYR-1 WLDGRRDVRVNTLAGYRHSLKPVIDNLGGLGLQQLRTS---DIDALVTLR
MAV_0253|M.avium_104 WLAAQR-VRTKAMTGYVTALRPLVDHLGGKAVQAVTKA---DIEKVVAAL
MSMEG_5754|M.smegmatis_MC2_155 WLLGQR-IRPKTMSAYITSLRPIVDHLGKRTVQSITKD---DLEQVVQAL
MLBr_01365|M.leprae_Br4923 YLIIERSIAANTLSSYRRDLIRYSKHLSDRGIEDLAKVGEHDVSEFLVAL
TH_1542|M.thermoresistible__bu HLSIERGVAANTLSSYRRDLRRYAEHLLARGIDDLAKVHESDVADFLVAL
MAB_1816c|M.abscessus_ATCC_199 RNFLG---------------------------------------------
MMAR_3607|M.marinum_M KKFIG---------------------------------------------
MUL_3581|M.ulcerans_Agy99 --------------------------------------------------
Mflv_2786|M.gilvum_PYR-GCK LNGTPISQRERRGRRAAEVLAYLRSRSGGAQYADVRDELGNPGVKALDRL
Mvan_3747|M.vanbaalenii_PYR-1 LNGTPISQRERRGRRAAEVLAYLRSRSGGAQYADVRDELGNPGVKALDRL
MAV_0253|M.avium_104 RGGT----------------------------------------------
MSMEG_5754|M.smegmatis_MC2_155 RAGT----------------------------------------------
MLBr_01365|M.leprae_Br4923 RRGD----------------------------------------------
TH_1542|M.thermoresistible__bu RRGD----------------------------------------------
MAB_1816c|M.abscessus_ATCC_199 --------GVRVSEAHAARMDAALRSMRTAHGTTMARQAKTILQGGLQLA
MMAR_3607|M.marinum_M --------GLRVSEATPARMDATLRSMRSAHGATMARQSKSILRGALQLA
MUL_3581|M.ulcerans_Agy99 --------------------------------------------------
Mflv_2786|M.gilvum_PYR-GCK VASGEVIRPSRGRYVAVRDSDPVQSKIPGNVSSRTVVTMLVVLSSALDDA
Mvan_3747|M.vanbaalenii_PYR-1 VASGEVIRPSRGRYVAVRDSDPVQSKIPGNVSSRTVVTMLVVLSSALDDA
MAV_0253|M.avium_104 --------SKMGTWNAPEKIK--GKQVRAKWGPSGTNHFIRRSRAVFEDL
MSMEG_5754|M.smegmatis_MC2_155 --------SKMGTWNAPTKLTKAAKKVRSPWAASSINPMLARTRSIFEDL
MLBr_01365|M.leprae_Br4923 --------PDSG---------------VAALSAVSAARALIAVRGLHRFA
TH_1542|M.thermoresistible__bu --------PDTG---------------AAGLSAVSAARALIAVRGLHRFA
MAB_1816c|M.abscessus_ATCC_199 VMANVIPANPVRDVQSLRSKTRPKGATALSADQLRTLLAEIRASEYCAGN
MMAR_3607|M.marinum_M VMASVLGANPVRDVQPLRSQSQPKGAVALTADQLRELLARLQTSDFCKEH
MUL_3581|M.ulcerans_Agy99 -MASTLGANPVRDVQPIRLKRRPTGATALSADDLHDLLVRLRADDYCQRN
Mflv_2786|M.gilvum_PYR-GCK MREGLVARNVARLVKRPA--VEHHEMATWTAEQAVRFREHVRGDRLAACW
Mvan_3747|M.vanbaalenii_PYR-1 MREGLVARNVARLVKRPA--VEHHEMATWTAEQAVRFREHVRGDRLAACW
MAV_0253|M.avium_104 VQQGVLGRNPAALVQPVP--MARAELNTLTAAQAERLLAATRNDVFAAAW
MSMEG_5754|M.smegmatis_MC2_155 VNQGVVVRNVAALVKPLAT-DTEKTMNTLNAEQVTQLLAVTDSHCYGIAW
MLBr_01365|M.leprae_Br4923 VAEGLVDLDVARAVRPPT--PGRRLPKSLTVDEVLALLESVGGE----SR
TH_1542|M.thermoresistible__bu AAEGVTELDVARSVKPPT--PSRRLPKSLTVDEVLALLAGAGGD----SE
. : . *: : .. : :
MAB_1816c|M.abscessus_ATCC_199 DLADPLIILMATGLRRSELLALRWSDFDANAGTLTVTGKLVRATGAGLQR
MMAR_3607|M.marinum_M DLIDPITLLIATGLRRSELLGLRWVDFDEKASMLAITGKVIRVPGEGLLR
MUL_3581|M.ulcerans_Agy99 DLVDPITVLIATGVRRSELLALRWTDFDESKQTIAATGNVVRV-------
Mflv_2786|M.gilvum_PYR-GCK LLT-------LAGLRRSEVLGLRWSDVDLDGGTVTIAQGRVVAEGQGTIT
Mvan_3747|M.vanbaalenii_PYR-1 LLT-------LAGLRRSEVLGLRWSDVDLDGGTVTIAQGRVVAEGQGTIT
MAV_0253|M.avium_104 RLA-------LMGLRRGEILGLSWDSVDLDAGLLTVRANRAPVPG-GVEV
MSMEG_5754|M.smegmatis_MC2_155 RLA-------VLGLRRGEILALRWDAVDFDAGTLAISAARLATAG-GATT
MLBr_01365|M.leprae_Br4923 ADG-------PLVLRNRALLELLYSTGSRISEAVGLDVDDVDTQARTVLL
TH_1542|M.thermoresistible__bu SDG-------PLTLRNRALLELLYSTGARISEAVGLDIDDVDTVARSVLL
:*. :* * : : .
MAB_1816c|M.abscessus_ATCC_199 IDEAKTAAGMRTVALPSFAVTALRHRQGRDYVG---------QQLMIFPS
MMAR_3607|M.marinum_M VDETKSAAGRRTIPLPKFAVDVLGKRRQLPYLG---------EQVVIFPS
MUL_3581|M.ulcerans_Agy99 --------------LP----GLLGSRRVPAVDG---------TTAVISPM
Mflv_2786|M.gilvum_PYR-GCK GD-PKSKRSRRALPMPAEVLAALRAFRLRQSEERLAIGSEYPDTGLVAVN
Mvan_3747|M.vanbaalenii_PYR-1 GD-PKSKRSRRALPMPAEVLAALRVFRLRQSEERLAIGSEYPDTGLVAVN
MAV_0253|M.avium_104 GS-PKTAAGLRTLPMPAELTVAMKQARKQQRERRLALGSKWVESGLVVVG
MSMEG_5754|M.smegmatis_MC2_155 GA-PKTKTSVRTLPMPEDLTAALRRVRKRQKEYRLKLGSKWTDSGYVVVD
MLBr_01365|M.leprae_Br4923 QG-KGGKQ--RLVPVGRPAVQALDAYLVRGRSDLARRGPGMLATPAIFLN
TH_1542|M.thermoresistible__bu RG-KGGKQ--RLVPIGRPAVSALDAYLVRGRPELVRRGRG---TPAIFLN
: : :
MAB_1816c|M.abscessus_ATCC_199 TAGTWRDPNNFGKQWRAVRDDLGVPG-VTTHSFRKTMATLIDDNGLSARV
MMAR_3607|M.marinum_M TAGTLRDPNNFGKEWRTAREELGVAE-VTTHSFRKTVATLIDDEGLSARI
MUL_3581|M.ulcerans_Agy99 HG------------------------------------------------
Mflv_2786|M.gilvum_PYR-GCK VIGLPIRPETYSGEFMRHAKDAGVPL-IRLHDVRHTAATMLLDRGTTPSA
Mvan_3747|M.vanbaalenii_PYR-1 VIGLPIRPETYSGEFMRHAKDAGVPL-IRLHDVRHTAATMLLDRGTTPSA
MAV_0253|M.avium_104 EDGAPPHPDTLSHVWAETIAAHKLPA-VRLHDARHTCATLMHQDGVPLAV
MSMEG_5754|M.smegmatis_MC2_155 EFGVAPYPDTLTAAWRKTLANAGLPH-VRLHDARHSCATLMHLSGVPTVV
MLBr_01365|M.leprae_Br4923 ARGGRLSRQSAWQVLQDAAEHAGITSGVSPHMLRHSFATHLLEGGADIRV
TH_1542|M.thermoresistible__bu ARGGRLSRQSAWQVLQDAAERAGIKAGVSPHTLRHSFATHLLDGGADVRV
MAB_1816c|M.abscessus_ATCC_199 GADQLGHAKVSMTQDRYMARGKVHHEVADLLD--ATISDE------
MMAR_3607|M.marinum_M GADHLGHSHVSMTQDRYMTRGRIHTQVAHLLDRAVRINDE------
MUL_3581|M.ulcerans_Agy99 ----------------------------------------------
Mflv_2786|M.gilvum_PYR-GCK TAKWLGH-DPAITLRVYGHVYDGALAAAGDTLLRGH----------
Mvan_3747|M.vanbaalenii_PYR-1 TAKWLGH-DPAITLRVYGHVYDGALAAAGDTLLRGH----------
MAV_0253|M.avium_104 ISAWLGHTNAAFTLATYGHSSDDSLAAAAQRLDALTGGKATPAEAK
MSMEG_5754|M.smegmatis_MC2_155 IAAWLGHQDPGFTLRTYAHSNHDALADAAAMLGSITTGKKK--EAK
MLBr_01365|M.leprae_Br4923 VQELMGH-ASVTTTQIYTLVTVQALREVWAGAHPRAK---------
TH_1542|M.thermoresistible__bu VQELLGH-ASVTTTQIYTMVTVHALREVWAGAHPRAR---------