For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MDNIRGKTIAITGAARGIGYATAEALLAHGARVVIGDRDVALQESSVAKLTNLGPVSGYPLDVTDQESFA TFLDKARTDGGGHIDVLINNAGVMPVGPFLDETEQSIRSSLEVNVYGVLTGCRLALPDMVKRRSGHIINI ASLSGVIPLPGQVVYVGAKYAVVGLTTALADEMAPHGVNVSVIMPPFTNTELIAGTKSGGAIKPVEPEDI AAAIVKTLNKPKTHVSVPPPLRFTAQAAQMLPPKGRRAMNKALGLDTVFLDVDTDKRKGYEERARAARGV VEGSQG
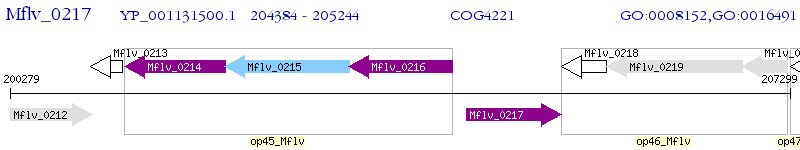
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0217 | - | - | 100% (286) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_3370 | - | 9e-28 | 37.68% (207) | short-chain dehydrogenase/reductase SDR |
| M. gilvum PYR-GCK | Mflv_2937 | - | 3e-26 | 32.42% (256) | short chain dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1896c | - | 2e-53 | 46.01% (276) | short chain dehydrogenase |
| M. tuberculosis H37Rv | Rv1865c | - | 2e-53 | 46.01% (276) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_01094 | - | 1e-26 | 31.02% (245) | short chain alcohol dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3405 | - | 4e-88 | 58.87% (282) | short chain dehydrogenase |
| M. marinum M | MMAR_2743 | - | 1e-51 | 44.04% (277) | short-chain type dehydrogenase |
| M. avium 104 | MAV_2700 | - | 1e-61 | 47.10% (276) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_0771 | - | 1e-141 | 87.72% (285) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_3427 | - | 1e-128 | 80.21% (283) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_3011 | - | 1e-51 | 43.88% (278) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_0690 | - | 1e-137 | 83.16% (285) | short chain dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1896c|M.bovis_AF2122/97 --MPGRTSIGVKIRDKVQDKVIAITGGARGIGLATAAALHNLGAKVAIGD
Rv1865c|M.tuberculosis_H37Rv --MPGRTSIGVKIRDKVQDKVIAITGGARGIGLATAAALHNLGAKVAIGD
MMAR_2743|M.marinum_M --MADSTTIGVRVR----DKVIVITGGARGIGLATATALHKLGAKVAIGD
MUL_3011|M.ulcerans_Agy99 --MADSTTIGVRVR----NKVIVITGGARGIGLATATALHKLGAKVAIGD
MAV_2700|M.avium_104 ---MDLT-----------DKVVAITGGARGIGLATAKAVLAEGAKVALGD
Mflv_0217|M.gilvum_PYR-GCK ---MDNIR----------GKTIAITGAARGIGYATAEALLAHGARVVIGD
MSMEG_0771|M.smegmatis_MC2_155 ---MDNIR----------GKTIAITGAARGIGYATAKALLAHGARVVIGD
Mvan_0690|M.vanbaalenii_PYR-1 ---MDNIR----------GKTIAITGAARGIGYATAEALLRRGARVVIGD
TH_3427|M.thermoresistible__bu ---MDNIR----------GKTIAITGAARGIGYATATALLARGARVVIGD
MAB_3405|M.abscessus_ATCC_1997 ---MDKIT----------GKNIAITGAARGIGYATATALLRRGARVVIGD
MLBr_01094|M.leprae_Br4923 MEGFA-------------GKVAVVTGAGSGIGQALAIELARSGAKLAISD
.* .:**.. *** * * : **::.:.*
Mb1896c|M.bovis_AF2122/97 IDEAMAKESGADLD---LDMYG-KLDVTDPDSFSGFLDAVER-QLGPIDV
Rv1865c|M.tuberculosis_H37Rv IDEAMAKESGADLD---LDMYG-KLDVTDPDSFSGFLDAVER-QLGPIDV
MMAR_2743|M.marinum_M VDEPAVKEAGADLG---LEVYG-KLDVTDPNSFSDFLDQVER-QLGPLDV
MUL_3011|M.ulcerans_Agy99 VGEPAVKEAGADLG---LEVYG-KLDVTDPNSFSDFLDQVER-QLGPLDV
MAV_2700|M.avium_104 LDTELAEKQAVELGGD-PAVVGLPLDVSDPASFAAFLDDVEA-RLGRLDV
Mflv_0217|M.gilvum_PYR-GCK RDVALQESSVAKLTNL-GPVSGYPLDVTDQESFATFLDKARTDGGGHIDV
MSMEG_0771|M.smegmatis_MC2_155 RDVALQESAVVELSKL-GQVSGYPLDVTDRESFATFLDKARTDGGGHIDV
Mvan_0690|M.vanbaalenii_PYR-1 RDVALQESAVAQLTKL-GPVSGYPLDVTDRDSFATFLDKARTDGGGHVDV
TH_3427|M.thermoresistible__bu RDVALQESAVAQLGRL-GPVSGYPLDVTDRESFATFLDKARTDGGGHIDV
MAB_3405|M.abscessus_ATCC_1997 RDVEALGSAVEGLKEE-GNVDAHPLDVTDPASFKRFLEAAS--AGGPIDV
MLBr_01094|M.leprae_Br4923 VDGEGLAQTEGQLKAIGASARTDRLDVTEREAFLTYADVVHE-NFGKVNQ
. . * ***:: :* : : . * ::
Mb1896c|M.bovis_AF2122/97 LVNNAGIMPVGRIVDEPDPVTRRILDINVYGVILGSKLAAQRMVPRGRGH
Rv1865c|M.tuberculosis_H37Rv LVNNAGIMPVGRIVDEPDPVTRRILDINVYGVILGSKLAAQRMVPRGRGH
MMAR_2743|M.marinum_M LVNNAGIMPVGRIVDEPDSVTRRILDINVYGVMVGSKLAAQRMVPRGRGH
MUL_3011|M.ulcerans_Agy99 LVNNAGIMPVGRIVDEPDSVTRRILDINVYGVMVGSKLAAQRMVPRGRGH
MAV_2700|M.avium_104 LVNNAGIMPTGPFVDESPTMSRRMIDVNVYGVLNGSRLAAARFVPRGAGH
Mflv_0217|M.gilvum_PYR-GCK LINNAGVMPVGPFLDETEQSIRSSLEVNVYGVLTGCRLALPDMVKRRSGH
MSMEG_0771|M.smegmatis_MC2_155 LINNAGVMPVGPFLEQSEQSIRSNIEVNVYGVLTGCQLALPDMVKRRRGH
Mvan_0690|M.vanbaalenii_PYR-1 LINNAGVMPIGPFLDQSEQSIRSSIEVNLYGVITGCQLALPDMIARRRGH
TH_3427|M.thermoresistible__bu LINNAGVMPIGPFLEQSEQAIRSSIEVNLYGVLTGCQLALPDMVRRRSGH
MAB_3405|M.abscessus_ATCC_1997 LVNNAGVMPIGPFLSTSDRAIRSMLEVNLYGVINGCQLALPEMVARRRGQ
MLBr_01094|M.leprae_Br4923 IYNNAGIAFTGDVEVSHFKDIERVMDVDYWGVVNGTKAFLPYLISSGDGH
: ****: * . . :::: :**: * : :: *:
Mb1896c|M.bovis_AF2122/97 VINVASLAGEIYAVGVATYCASKHAVVAFTDSARLEYRSAG--VKFSMVL
Rv1865c|M.tuberculosis_H37Rv VINVASLAGEIYAVGVATYCASKHAVVAFTDSARLEYRSAG--VKFSMVL
MMAR_2743|M.marinum_M VINVASLAGEIYVVGLATYCASKHAVIAFTDAARIEYRSTG--VKFSMVL
MUL_3011|M.ulcerans_Agy99 VINVASLAGEIYVVGLATYCASKHAVIAFTDAARIEYRTTG--VKFSMVL
MAV_2700|M.avium_104 IVNIASLAGVTGEPGMATYCGTKHFVVGFTESLHRELRPHR--VGVSLVL
Mflv_0217|M.gilvum_PYR-GCK IINIASLSGVIPLPGQVVYVGAKYAVVGLTTALADEMAPHG--VNVSVIM
MSMEG_0771|M.smegmatis_MC2_155 IINIASLSGVIPLPGQVVYVGAKYAVVGLSTALADEMAPYG--VDVSVIM
Mvan_0690|M.vanbaalenii_PYR-1 IINIASLSGLIPVPGQVVYVGAKFGVVGLSTALADEVAPQG--VDVSVIM
TH_3427|M.thermoresistible__bu IINIASMSGLIPVPGQVPYNAAKFGVVGLSVALADEMAPYG--VEVSVVM
MAB_3405|M.abscessus_ATCC_1997 VVNIASVAGLMAVPGMAVYNASKFGVVGLSRALSDEFAPQG--VQVSAVL
MLBr_01094|M.leprae_Br4923 VINISSVFGLFSVPGQAAYNSAKFAVRGFTEALREEMALAGRPVNVTTVY
::*::*: * * . * .:*. * .:: : * * .: :
Mb1896c|M.bovis_AF2122/97 PSFVNTELIAGTGG---IKGFKNAEPADIADAIVGLIVHPKPRVRVTKAA
Rv1865c|M.tuberculosis_H37Rv PSFVNTELIAGTGG---IKGFKNAEPADIADAIVGLIVHPKPRVRVTKAA
MMAR_2743|M.marinum_M PTFVNTELASGTPG---MKGFKNAEPSDIADAIVALVANPKPRVRVTKAA
MUL_3011|M.ulcerans_Agy99 PTFVNTELASGTPG---MKGFKNAEPSDIADAIVALVANPKPRVRVTKAA
MAV_2700|M.avium_104 PGIINTELSAGTKVPGWARPLATAEPEDVAAGIVAAVSKDKPRKTVPATL
Mflv_0217|M.gilvum_PYR-GCK PPFTNTELIAGTKS---GGAIKPVEPEDIAAAIVKTLNKPKTHVSVPPPL
MSMEG_0771|M.smegmatis_MC2_155 PPFTNTDLISGTKS---GGAIKPVEPEEIAAAIVKTLNKPKTHVSVPPPL
Mvan_0690|M.vanbaalenii_PYR-1 PPFTNTDLISGTTS---SGALKPVEPEDIAAAVVKTLNKPKTHVSVPPPL
TH_3427|M.thermoresistible__bu PPFTRTELISGTKE---TAATKPAEPEDIAAAIVKTLDKPKTHVSVPGPL
MAB_3405|M.abscessus_ATCC_1997 PTFTNTELIAGTDS---TGAQKPVEPEDIAAAVVRIIEKPRVQVTVPKPL
MLBr_01094|M.leprae_Br4923 PGGIKTAIARNATA---AEGLDVSKIASRFDTWVAHTSPQHAARIILKAV
* .* : .: : . * : : .
Mb1896c|M.bovis_AF2122/97 GSMIVAQRFMPRQVSEGLNRLLGGEHVFTDDVDMEKRRTYEARARGEE--
Rv1865c|M.tuberculosis_H37Rv GSMIVAQRFMPRQVSEGLNRLLGGEHVFTDDVDMEKRRTYEARARGEE--
MMAR_2743|M.marinum_M GAMIASQKYLPRWLSEGLNRLLGGEHVFTDDVDAAKREAYEARARGETGE
MUL_3011|M.ulcerans_Agy99 GAMIASQKYLPRWLSEGLNRLLGGEHVFTDDVDAAKREAYEARARGETGE
MAV_2700|M.avium_104 GALLKSVSLLPDTPRFAVAHAVRFDRLVSG-ADPDARAAYHRRLAEES--
Mflv_0217|M.gilvum_PYR-GCK RFTAQAAQMLPPKGRRAMNKALGLDTVFLD-VDTDKRKGYEERARAARGV
MSMEG_0771|M.smegmatis_MC2_155 RFTAQAAQMLPPKGRRWLNKKLGLDNVFLE-FDTAKRKAYEDRAQAALGV
Mvan_0690|M.vanbaalenii_PYR-1 RFTAQAAQMLPPRGRRWLSRKLGLDRVFLE-FDTTKRQGYEQRAQSALGV
TH_3427|M.thermoresistible__bu RFIAQAAQTLGPRGRRWLNKRLGLDTVFLE-FDQQARQGYEQRAQQALGV
MAB_3405|M.abscessus_ATCC_1997 GAVTTIVNSLPTGVRRFMSKKTGTDRVFLD-FDTSARTAYENRAGSSLGV
MLBr_01094|M.leprae_Br4923 RKKKARVLVGPDAKVANVVVRFSGGAGYQRLFAQVASRLILNQR------
: :
Mb1896c|M.bovis_AF2122/97 ---------------
Rv1865c|M.tuberculosis_H37Rv ---------------
MMAR_2743|M.marinum_M S--------------
MUL_3011|M.ulcerans_Agy99 S--------------
MAV_2700|M.avium_104 ---------------
Mflv_0217|M.gilvum_PYR-GCK VEGSQG---------
MSMEG_0771|M.smegmatis_MC2_155 VESEKK---------
Mvan_0690|M.vanbaalenii_PYR-1 VEGSENKPSPKM---
TH_3427|M.thermoresistible__bu LEGGNKQQDRSGQKS
MAB_3405|M.abscessus_ATCC_1997 TKPDGE---------
MLBr_01094|M.leprae_Br4923 ---------------