For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RNIPLADVDHLPHEVLPISTDYADGQLLDWHRHRRAQFLYATTGTMIVDTDAGTWTVPTHRAVMIPARTR HRVRMFEVRTGSLYVEPSAAPWWPTSCVVVDVSPLMHELLSAAADLDVDYDRSGRGGALITLLLHEIASF TALPLHVDIPWSADLAELCRTYLDAPDVSVTNAEWARRLRTSERALTRRFRDEVAMSPAAWRVRARLLAA VPLLRDHSVTQVAGRLGYATPAAFSYAFGRAFNIAPSSVR*
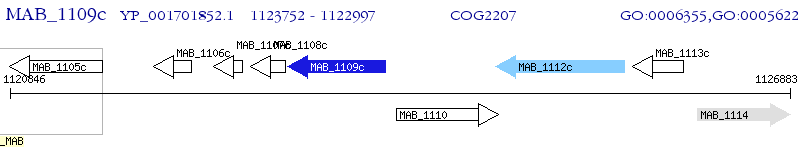
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1109c | - | - | 100% (251) | AraC family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_1337c | - | 5e-25 | 32.75% (229) | AraC family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_0557 | - | 2e-08 | 33.33% (96) | AraC family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3863 | - | 3e-25 | 31.51% (238) | AraC family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_2370 | - | 9e-13 | 27.27% (242) | AraC family transcriptional regulator |
| M. tuberculosis H37Rv | Rv3833 | - | 4e-25 | 31.51% (238) | AraC family transcriptional regulator |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_0193 | - | 8e-24 | 33.33% (222) | transcriptional regulator, AraC family protein |
| M. smegmatis MC2 155 | MSMEG_4601 | - | 2e-59 | 48.13% (241) | transcriptional regulator, AraC family protein |
| M. thermoresistible (build 8) | TH_2340 | - | 3e-16 | 27.51% (229) | transcriptional regulatory protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_4276 | - | 1e-13 | 25.42% (236) | AraC family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3863|M.bovis_AF2122/97 --------------------------MSENSHHRLATTSLTLPPGARIER
Rv3833|M.tuberculosis_H37Rv --------------------------MSENSHHRLATTSLTLPPGARIER
MAV_0193|M.avium_104 --------------------------MSEIGQHGLATSAQTLPPGARIAR
MAB_1109c|M.abscessus_ATCC_199 -------------------MRNIPLADVDHLPHEVLPISTDYADGQLLDW
MSMEG_4601|M.smegmatis_MC2_155 -------------------MR-TTVETHDNLARDVLALATDYSYGEVLAP
Mflv_2370|M.gilvum_PYR-GCK MTVSASSDRVLSETANL---SPRRVIDLRRGGRALGGSYLYEGDELITGW
Mvan_4276|M.vanbaalenii_PYR-1 MTVSASSDRILSEAANLGAGPPRRVIDLRRGGRALGGSYLYEGDALITGW
TH_2340|M.thermoresistible__bu VTVSALSARNLASSAKP------PQIDLRRGGRALGGSYLYEGDKLQTGW
: :
Mb3863|M.bovis_AF2122/97 HRHPSHQIVYPSAGAVSVTTHAGTWITPVNRAIWIPAGCWHQHKFHGHTQ
Rv3833|M.tuberculosis_H37Rv HRHPSHQIVYPSAGAVSVTTHAGTWITPVNRAIWIPAGCWHQHKFHGHTQ
MAV_0193|M.avium_104 HRHPLHQIVYPSTGAVSVTTPAGTWITPANRAIWIPAGCWHEHKFHGHTN
MAB_1109c|M.abscessus_ATCC_199 HRHRRAQFLYATTGTMIVDTDAGTWTVPTHRAVMIPARTRHRVRMFEVRT
MSMEG_4601|M.smegmatis_MC2_155 HRHRRAQFLYGVGGTMDVRTEHGSWLVPTERAVLIPPQTQHAVTMWGVST
Mflv_2370|M.gilvum_PYR-GCK HSHDVHQIEYALHGVVEVETDSAHYLLPPQQAAWIPAGLEHQAVMNPDVK
Mvan_4276|M.vanbaalenii_PYR-1 HSHDVHQIEYALHGVVEVETDSAHYLLPPQQAAWIPAGLEHQAVMNPDVK
TH_2340|M.thermoresistible__bu HSHDVHQIEYAIGGVVEVQTRTGHYLLPPQQAAWLPVGLEHSATMNPDVK
* * *: * *.: * * . : * .:* :* * :
Mb3863|M.bovis_AF2122/97 FHGVALDPQRYRGGPATPTVLAVNPLMRELIIACSQADRT-DTDEH----
Rv3833|M.tuberculosis_H37Rv FHGVALDPQRYRGGPATPTVLAVNPLMRELVIACSQADRT-DTDEH----
MAV_0193|M.avium_104 FHGVALDPARYRRGPAAPAVLAVTPLMRELIIACSRARDT-DAGAH----
MAB_1109c|M.abscessus_ATCC_199 GS-LYVEPSAAPWWPTSCVVVDVSPLMHELLSAAADLDVDYDRSGR----
MSMEG_4601|M.smegmatis_MC2_155 RS-LYIEPGAVPWFPTRCTVVDVPTLLRELLVAAVAVPLDRPPDAR----
Mflv_2370|M.gilvum_PYR-GCK TVAVMFDPALIADGGDRARILAVSPLIREMMIYALRWPIDRGPADAASDD
Mvan_4276|M.vanbaalenii_PYR-1 TVAVMFDPTLIPDGGDRARILAVSPLIREMMIYALRWHIERGPG-AGDDD
TH_2340|M.thermoresistible__bu TVAVMFDPQLVPDAGNRARILAVSPLIREMMIYALRWPIDRPHG-----D
: .:* :: * .*::*:: .
Mb3863|M.bovis_AF2122/97 --HRMLAVLQDQLPTTSIREPLWVPSPT-DRRLRHACALIADNLTQPLTL
Rv3833|M.tuberculosis_H37Rv --HRMLAVLQDQLPTTSIREPLWVPSPT-DRRLRHACALIADNLTQPLTL
MAV_0193|M.avium_104 --HRMLAVLHDQLQATSVAEPLWIPTAV-DGRLRAACALIADNLREPLTL
MAB_1109c|M.abscessus_ATCC_199 --GGALITLLLHEIASFTALPLHVDIPW-SADLAELCRTYLDAPDVSVTN
MSMEG_4601|M.smegmatis_MC2_155 --DAALFALTLHEVAGCTSLPLDLPLPQ-PGALSEQCRRFLAAPRIDTTT
Mflv_2370|M.gilvum_PYR-GCK TVADGFFRTMAALVCEALDHEAPLSLPTSDHPIVAAALDYTKRHLDTATA
Mvan_4276|M.vanbaalenii_PYR-1 GVADGFFRTLAALVCEALDHEAPLSLPTSDNPIVAAAMAYTKQHLDTVTC
TH_2340|M.thermoresistible__bu EISDTFFRTLAHLVSEALDHEAPLSLPTSDHPIVAAAMAYTKEHLQSVTA
: : . : . *
Mb3863|M.bovis_AF2122/97 QQIGGRIGVSQRTLSRLFSDELGMTFPQWRTQLRLQHALVLLAER-HDVT
Rv3833|M.tuberculosis_H37Rv QQIGGRIGVSQRTLSRLFSDELGMTFPQWRTQLRLQHALVLLAER-HDVT
MAV_0193|M.avium_104 RQIGERIGVGQRTLSRLFRDELAMTFPQWRTQVRLQHALVLLAER-RDVT
MAB_1109c|M.abscessus_ATCC_199 AEWARRLRTSERALTRRFRDEVAMSPAAWRVRARLLAAVPLLRD--HSVT
MSMEG_4601|M.smegmatis_MC2_155 GWWANALHVSERTVARLFRKELSMTVAEWQRRACVLHSLRQLTQG-VAVS
Mflv_2370|M.gilvum_PYR-GCK EEVSRAVSVSERTLRRLFADTLGLPWRTYLLHARMLRAMAPLAAPAQSVQ
Mvan_4276|M.vanbaalenii_PYR-1 EEVSRAVSVSERTLRRLFAETLGLPWRTYLLHARMLRAMALLTAPTQSVQ
TH_2340|M.thermoresistible__bu EEVSRAVAVSERTLRRQFRQATGISWRTYLLHARMLKAMALLAAPDQSVQ
. : ..:*:: * * . .:. : : : :: * *
Mb3863|M.bovis_AF2122/97 SVASECGWATPSAFIDTYRQAFGHTPGQAAKPMAATRLTRLRRARDRR
Rv3833|M.tuberculosis_H37Rv SVASECGWATPSAFIDTYRQAFGHTPGQAAKPMAATRLTRLRRARDRR
MAV_0193|M.avium_104 SVAAECGWATPSAFIDTYRRAFGHTPGRLAP-----RPS---------
MAB_1109c|M.abscessus_ATCC_199 QVAGRLGYATPAAFSYAFGRAFNIAPSSVR------------------
MSMEG_4601|M.smegmatis_MC2_155 AVASDLGYSSPAAFSTMFSRALGTSPSAFSPRRLNL------------
Mflv_2370|M.gilvum_PYR-GCK ETATAVGFDSVSSFTRAFAQFCGETPSAYRKRVADSD-----------
Mvan_4276|M.vanbaalenii_PYR-1 GTATAVGFDSVSSFTRAFTQFCGETPSAYRKRVADASD----------
TH_2340|M.thermoresistible__bu ETASAVGFDSLSAFTRAFTQFCGETPSAYRHRISTDPHSRPTSADPIG
.* *: : ::* : : . :*.