For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
IRATFVAHTAAPSGAELATLRLLSAVRNSSTGLILEPSVVYTEDGPMVNAMKSQGISTSVLANTFDSRSV TIRGSGWLRRLAGATSLVRVGWSLGAAVREHGADVLVAGSTKALIMSAVAAKRARIPLIWHVHDRISAEY FGAALAFLIRILGKLVADGYIANSRSTESSLRPGRLPGVIAYPGLDFQHTPAVEHAPQRPAAETVIAVVG RLTQWKGQDVFLRALANTRVRPQRVYLVGGTFFGEESFRTELEDLASELQLPVTFTGHLDDPTDILSIAD ILVHCSVIAEPFGQVVVEGLRAGCAVIATQPGGPAETIESGVHGLLVRAGDTGQLTDALDHLIEDRDLRI RLAQAGRDRARRFDIADSAQTVGTFLSDMLARRSTTPLAGRRV*
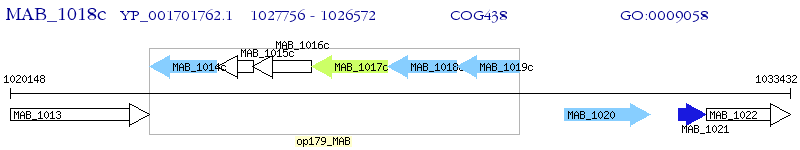
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1018c | - | - | 100% (394) | glycosyl transferase |
| M. abscessus ATCC 19977 | MAB_2894c | - | 1e-10 | 29.84% (191) | phosphatidylinositol alpha-mannosyltransferase |
| M. abscessus ATCC 19977 | MAB_1014c | - | 1e-09 | 27.80% (277) | glycosyl transferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0496 | - | 3e-12 | 32.18% (174) | mannosyltransferase |
| M. gilvum PYR-GCK | Mflv_1500 | - | 3e-12 | 26.16% (367) | glycosyl transferase, group 1 |
| M. tuberculosis H37Rv | Rv0486 | - | 4e-12 | 32.18% (174) | mannosyltransferase |
| M. leprae Br4923 | MLBr_00452 | - | 5e-11 | 30.17% (179) | putative glycosyltransferase |
| M. marinum M | MMAR_2092 | pimA | 1e-10 | 32.03% (153) | alpha-mannosyltransferase PimA |
| M. avium 104 | MAV_4082 | - | 2e-12 | 35.98% (164) | transferase |
| M. smegmatis MC2 155 | MSMEG_0933 | - | 1e-10 | 29.39% (228) | hypothetical protein MSMEG_0933 |
| M. thermoresistible (build 8) | TH_3188 | - | 5e-13 | 36.00% (175) | PUTATIVE glycosyl transferase, group 1 |
| M. ulcerans Agy99 | MUL_3252 | pimA | 1e-10 | 32.03% (153) | alpha-mannosyltransferase PimA |
| M. vanbaalenii PYR-1 | Mvan_5246 | - | 5e-14 | 25.68% (366) | glycosyl transferase, group 1 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2092|M.marinum_M --------------------------------------------------
MUL_3252|M.ulcerans_Agy99 ---------------------------MAHVSATMVGRLVRCPPRAAAGA
MLBr_00452|M.leprae_Br4923 --------------------------------------------------
Mb0496|M.bovis_AF2122/97 MAGVRHDDGSGLIAQRRPVRGEGATRSRGPSGPSNRNVSAADDPRRVALL
Rv0486|M.tuberculosis_H37Rv MAGVRHDDGSGLIAQRRPVRGEGATRSRGPSGPSNRNVSAADDPRRVALL
MSMEG_0933|M.smegmatis_MC2_155 ----------------------------------MRLATDLETPRRVAVL
MAV_4082|M.avium_104 --------------------------------------------MRIAMI
TH_3188|M.thermoresistible__bu ------------------------------------------------MV
Mflv_1500|M.gilvum_PYR-GCK -------------------------------------------------M
Mvan_5246|M.vanbaalenii_PYR-1 -------------------------------------------------M
MAB_1018c|M.abscessus_ATCC_199 -----------------------------------------------MIR
MMAR_2092|M.marinum_M -MRIGMVCPYSFDVAGGVQSHVLQLAEVMRARGHEVSVLAPASPHAALPD
MUL_3252|M.ulcerans_Agy99 LMRIGMVCPYSFDVAGGVQSHVLQLAEVMRARGHEVSVLAPASPHAALPD
MLBr_00452|M.leprae_Br4923 -MRIGMICPYSFDVPGGVQSHVLQLAEVMRARGQQVRVLAPASPDVSLPE
Mb0496|M.bovis_AF2122/97 AVHTSPLAQPGTGDAGGMNVYMLQSALHLARRGIEVEIFTRATASADPPV
Rv0486|M.tuberculosis_H37Rv AVHTSPLAQPGTGDAGGMNVYMLQSALHLARRGIEVEIFTRATASADPPV
MSMEG_0933|M.smegmatis_MC2_155 SVHTSPLAQPGTGDAGGMNVYVLQTALQLARRGVEVEVFTRATSSADAPV
MAV_4082|M.avium_104 SEHASPLAHLGGVDAGGQNVHVAELSCALARRGHDVVVYTRRDDPRLPDS
TH_3188|M.thermoresistible__bu SEHASPLAALGGVDAGGQNVHVAELSAALTRRGHEVTVYTRRDDRGLPDR
Mflv_1500|M.gilvum_PYR-GCK TGGVSALVLIDAFRMGGAETLLAPMIVASRDTDVSMDVVSISPSEWNSEK
Mvan_5246|M.vanbaalenii_PYR-1 TRGISALVLIDAFRMGGAETLLAPMIVASRDTDVTMDVVSISPADWNSEK
MAB_1018c|M.abscessus_ATCC_199 ATFVAHTAAPSGAELATLRLLSAVRNSSTGLILEPSVVYTEDGPMVNAMK
. . . . : :
MMAR_2092|M.marinum_M YVVSGGKAVPIPYNGSVARLR-------FGPATHRKVKKWLSEGD-----
MUL_3252|M.ulcerans_Agy99 YVVSGGKAVPIPYNGSVARLR-------FGPATHRKVKKWLSEGD-----
MLBr_00452|M.leprae_Br4923 YVVSAGRAIPIPYNGSVARLQ-------FSPAVHSRVRRWLVDGD-----
Mb0496|M.bovis_AF2122/97 VRVAPGVLVRNVVAGPFEGLDKYDLPTQLCAFAAGVLRAEAVHEPG---Y
Rv0486|M.tuberculosis_H37Rv VRVAPGVLVRNVVAGPFEGLDKYDLPTQLCAFAAGVLRAEAVHEPG---Y
MSMEG_0933|M.smegmatis_MC2_155 VPVAPGVLVRNVVAGPFEGLDKNDLPTQLCAFTAGVLRAEATHEPG---Y
MAV_4082|M.avium_104 VETPHGYSVVHVPAGPARVLPKDELLQYMYGFAQFLRQQWCSDGP-----
TH_3188|M.thermoresistible__bu VDTPLGYTVVHVPAGPAEQLPKDELLPYMGPFAQFLDAEWSAHRP-----
Mflv_1500|M.gilvum_PYR-GCK TMTILAEAGIPTRSLGIRRLLD--------PVALPRLVRTIRQGR-----
Mvan_5246|M.vanbaalenii_PYR-1 TMAILADAGIATRSLGIRRLLD--------PVALPRLVKTIRQGR-----
MAB_1018c|M.abscessus_ATCC_199 SQGISTSVLANTFDSRSVTIRGSGWLRRLAGATSLVRVGWSLGAAVREHG
: .
MMAR_2092|M.marinum_M FDVLHLHEPNAPSLSMLALNIAEGPIVATFHTSTTKS--------LTLTV
MUL_3252|M.ulcerans_Agy99 FDVLHLHEPNAPSLSMLALNIAEGPIVATFHTSTTKS--------LTLTV
MLBr_00452|M.leprae_Br4923 FDVLHLHEPNAPSLSMWALRVAEGPIVATFHTSTTKS--------LTLSV
Mb0496|M.bovis_AF2122/97 YDIVHSHYWLSGQVGWLARDRWAVPLVHTAHTLAAVKNAALADGDGPEPP
Rv0486|M.tuberculosis_H37Rv YDIVHSHYWLSGQVGWLARDRWAVPLVHTAHTLAAVKNAALADGDGPEPP
MSMEG_0933|M.smegmatis_MC2_155 YDVVHSHYWLSGQVGWLARDRWAVPLVHTAHTLAAVKNAALAAGDAPEPP
MAV_4082|M.avium_104 -DVAHAHFWMSGVATQQAARAHSIPTVQTFHALGLTKRLHQGSDD-TSPD
TH_3188|M.thermoresistible__bu -DVVHAHFWMSGIATQLASRHLDLPAVQTFHALGVVKKRHQGAED-TSPQ
Mflv_1500|M.gilvum_PYR-GCK YDVVHAHLEMAMTLAVPAAALAGRPAVCTFHHVARPLEG------RAAWR
Mvan_5246|M.vanbaalenii_PYR-1 YDIVHAHLEMAMTLAVPAAALVGRPAVCTFHHVARPLQG------RAAWR
MAB_1018c|M.abscessus_ATCC_199 ADVLVAGSTKALIMSAVAAKRARIPLIWHVHDR------------ISAEY
*: : * * : * .
MMAR_2092|M.marinum_M FQSILRPMHEKIVGRIAVSDLARRWQMEALGSD----AVEIPNGVDVGSF
MUL_3252|M.ulcerans_Agy99 FQSILRPMHEKIVGRIAVSDLARRWQMEALGSD----AVEIPNGVDVGSF
MLBr_00452|M.leprae_Br4923 FQGVLRPWHEKIIGRIAVSDLARRWQMEALGSD----AVEIPNGVNVDSL
Mb0496|M.bovis_AF2122/97 LRTVGEQQVVDEADRLIVNTDDEARQVISLHGADPARIDVVHPGVDLDVF
Rv0486|M.tuberculosis_H37Rv LRTVGEQQVVDEADRLIVNTDDEARQVISLHGADPARIDVVHPGVDLDVF
MSMEG_0933|M.smegmatis_MC2_155 LRAVGEQQVVDEADRLIVNTEVEAQQLVSLHNADRSRIDVVHPGVDLDVF
MAV_4082|M.avium_104 CRIDVEARVAREADWVAATSTDEVFELVRMGRA-RSRTSVVPCGVDLDAF
TH_3188|M.thermoresistible__bu ERLQLEAMVARGANWVAATCTDEVFELMRMGRS-RSRISVVPCGVDLDLF
Mflv_1500|M.gilvum_PYR-GCK ERLAVEAATRSRRALFVSEASRLSFAQNYRPKGMPANWEVMHNGIDIANF
Mvan_5246|M.vanbaalenii_PYR-1 ERLAVEAATRSRRALFVSEASRRSFAETYRPNRMPKNWEVMHNGIDISNF
MAB_1018c|M.abscessus_ATCC_199 FGAALAFLIRILGKLVADGYIANSRSTESSLRPGRLPGVIAYPGLDFQHT
. *::.
MMAR_2092|M.marinum_M ASSSCL-DGYPRPG-----KTVLFLGRYDEPRKGMSVLLEALPKLVERFS
MUL_3252|M.ulcerans_Agy99 ASSSCL-DGYPRPG-----KTVLFLGRYDEPRKGMSVLLEALPKLVERFI
MLBr_00452|M.leprae_Br4923 SSAPQL-AGYPRLG-----KTVLFLGRYDEPRKGMSVLLDALPGVMECFD
Mb0496|M.bovis_AF2122/97 RPGDRR-AARAALGLPVDERVVAFVGRIQ-PLKAPDIVLRAAAKLPG---
Rv0486|M.tuberculosis_H37Rv RPGDRR-AARAALGLPVDERVVAFVGRIQ-PLKAPDIVLRAAAKLPG---
MSMEG_0933|M.smegmatis_MC2_155 TPGSRD-AARAVFGLPTDQKIVAFVGRIQ-PLKAPDILLRAAAKLPG---
MAV_4082|M.avium_104 TPDGPV-AARGAR------PRIVTVGRLV-PRKGFDTIVRALPRIPD---
TH_3188|M.thermoresistible__bu TPDGPQ-APRGER------RRIVTVGRFV-PRKGFDTLVRALPAIPN---
Mflv_1500|M.gilvum_PYR-GCK APGQADPGVRSELAGRDGDSLVAVLPAAFRDFKGIPVAIKAWPAVIEKFP
Mvan_5246|M.vanbaalenii_PYR-1 SPGQADPRVRSELAAG-TQGPVVVLPAAFRDFKGIPVAIKAWPAVKQRFP
MAB_1018c|M.abscessus_ATCC_199 PAVEHAPQRPAAET------VIAVVGRLT-QWKGQDVFLRALANTRVR--
. : : *. : * .
MMAR_2092|M.marinum_M DVQLLIVGRGDEDELRDEAG-----------ELAEHMRFLGQVDDAGKAS
MUL_3252|M.ulcerans_Agy99 DVQLLIVGRGDEDELRDEAG-----------ELAEHMRFLGQVDDAGKAS
MLBr_00452|M.leprae_Br4923 DVQLLIVGRGDEEQLRSQAG-----------GLVEHIRFLGQVDDAGKAA
Mb0496|M.bovis_AF2122/97 -VRIIVAGGPSGSGLASPD---GLVRLADELGISARVTFLPPQSHTDLAT
Rv0486|M.tuberculosis_H37Rv -VRIIVAGGPSGSGLASPD---GLVRLADELGISARVTFLPPQSHTDLAT
MSMEG_0933|M.smegmatis_MC2_155 -VRVLIAGGPSGSGLAQPD---TLVRLADELGISDRVTFLPPQSREQLVN
MAV_4082|M.avium_104 -AELVIVGGPPAAELAGDEQARRLQRLAGELGVADRVRLLGGVTRAQMPA
TH_3188|M.thermoresistible__bu -AELVVVGGPRRSQLRTEPEARRLCTLAEELGVADRVQLYGSVARADMPA
Mflv_1500|M.gilvum_PYR-GCK SAVLALTGGGELESELR--------QLVADLGLGDSVNFVG--VRTDMPE
Mvan_5246|M.vanbaalenii_PYR-1 TAVLALTGGGELEGELR--------RLVADLGLGDSVNFVG--VRTDMPE
MAB_1018c|M.abscessus_ATCC_199 PQRVYLVGGTFFGEESFRT-------ELEDLASELQLPVTFTGHLDDPTD
: :.* : .
MMAR_2092|M.marinum_M AMRSADVYCAPNTGGESFGIVLVEAMAAGTPVVASDLDAFRRVLRDGEIG
MUL_3252|M.ulcerans_Agy99 AMRSADVYCAPNTGGESFGIVLVEAMAAGTPVVASDLDAFRRVLRDGEIG
MLBr_00452|M.leprae_Br4923 AMRSADVYCAPNIGGESFGIVLVEAMAAGTPVVASDLDAFRRVLRDGEVG
Mb0496|M.bovis_AF2122/97 LFRAADLVAVPSYS-ESFGLVAVEAQACGTPVVAAAVGGLPVAVRDGITG
Rv0486|M.tuberculosis_H37Rv LFRAADLVAVPSYS-ESFGLVAVEAQACGTPVVAAAVGGLPVAVRDGITG
MSMEG_0933|M.smegmatis_MC2_155 VYRAADLVAVPSYS-ESFGLVAVEAQACGTPVVAAAVGGLPVAVADGVSG
MAV_4082|M.avium_104 LLRSADVVACTPWY-EPFGIVPLEAMACGVPVVATAVGGIRDTVVDGATG
TH_3188|M.thermoresistible__bu VLRSADVVACTPWY-EPFGIVPLEAMACGVPVVAAAVGGMLDTVVHDVTG
Mflv_1500|M.gilvum_PYR-GCK VYRAADVVVLPSTHGENLPTVLIESSATACAVAASRVGGIPDIVLDGQTG
Mvan_5246|M.vanbaalenii_PYR-1 VYRAADVVVLPSTHGENLPTVLIEASATACAIAASRVGGIPDIVLDGATG
MAB_1018c|M.abscessus_ATCC_199 ILSIADILVHCSVIAEPFGQVVVEGLRAGCAVIATQPGGPAETIESGVHG
**: * : * :*. . .: *: .. : . *
MMAR_2092|M.marinum_M RLVPVDDSDALAAALIEVLDSQE--LRERYVAAGTEAVRRYDWSVVASQI
MUL_3252|M.ulcerans_Agy99 RLVPVDDSDALAAALIEVLDSQE--LRERYVAAGTEAVRRYDWSVVASQI
MLBr_00452|M.leprae_Br4923 HLVPAGDSAALADALVALLRNDV--LRERYVAAGAEAVRRYDWSVVASQI
Mb0496|M.bovis_AF2122/97 TLVSGHEVGQWADAIDHLLRLCAGPRGRVMSRAAARHAATFSWENTTDAL
Rv0486|M.tuberculosis_H37Rv TLVSGHEVGQWADAIDHLLRLCAGPRGRVMSRAAARHAATFSWENTTDAL
MSMEG_0933|M.smegmatis_MC2_155 ALVDGHDIGDWADTISEVLDREP----AALSRASAEHAAQFSWAHTVDAL
MAV_4082|M.avium_104 RLVPPKDPDALGEAVAALLRDGR-RGRTLGEAGRERARARYSWDRVAADT
TH_3188|M.thermoresistible__bu RLVPPRRPAELADTINQLLRDSF-LRQSLGAAGRDRARARYSWDRVATDT
Mflv_1500|M.gilvum_PYR-GCK LLFEAGSVEGFADTVCRLLGDAE-LRRRLGAAASERARSEFSATAWLARL
Mvan_5246|M.vanbaalenii_PYR-1 LLFEPGSADGLAEAVCRLLDDVE-LRRRMGAAASERARTEFSATAWLARL
MAB_1018c|M.abscessus_ATCC_199 LLVRAGDTGQLTDALDHLIEDRD--LRIRLAQAGRDRARRFDIADSAQTV
*. :: :: . :.
MMAR_2092|M.marinum_M MRVYETVAGAGVK---------------VQVAS----------
MUL_3252|M.ulcerans_Agy99 MRVYETVAGAGVK---------------VQVAS----------
MLBr_00452|M.leprae_Br4923 MRVYETVATSGSK---------------VQVAS----------
Mb0496|M.bovis_AF2122/97 LASYRRAIGEYNAERQRRGGEVISDLVAVGKPRHWTPRRGVGA
Rv0486|M.tuberculosis_H37Rv LASYRRAIGEYNAERQRRGGEVISDLVAVGKPRHWTPRRGVGA
MSMEG_0933|M.smegmatis_MC2_155 LASYSRAMSDYRARHPRP--------AARRSGRRFSMRRGVRT
MAV_4082|M.avium_104 ERIYERLSPARRG---------------VGTPSTRSG------
TH_3188|M.thermoresistible__bu QRIYDRLVPADYR---------------SATASQMSSG-----
Mflv_1500|M.gilvum_PYR-GCK RTTYLEAMGDRR-------------------------------
Mvan_5246|M.vanbaalenii_PYR-1 RTTYLEAMGVPR-------------------------------
MAB_1018c|M.abscessus_ATCC_199 GTFLSDMLARRST---------------TPLAGRRV-------