For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTGGVSALVLIDAFRMGGAETLLAPMIVASRDTDVSMDVVSISPSEWNSEKTMTILAEAGIPTRSLGIRR LLDPVALPRLVRTIRQGRYDVVHAHLEMAMTLAVPAAALAGRPAVCTFHHVARPLEGRAAWRERLAVEAA TRSRRALFVSEASRLSFAQNYRPKGMPANWEVMHNGIDIANFAPGQADPGVRSELAGRDGDSLVAVLPAA FRDFKGIPVAIKAWPAVIEKFPSAVLALTGGGELESELRQLVADLGLGDSVNFVGVRTDMPEVYRAADVV VLPSTHGENLPTVLIESSATACAVAASRVGGIPDIVLDGQTGLLFEAGSVEGFADTVCRLLGDAELRRRL GAAASERARSEFSATAWLARLRTTYLEAMGDRR
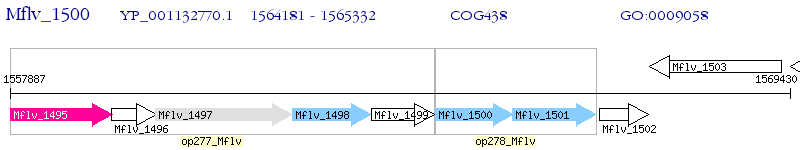
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1500 | - | - | 100% (383) | glycosyl transferase, group 1 |
| M. gilvum PYR-GCK | Mflv_3834 | - | 4e-22 | 28.46% (369) | phosphatidylinositol alpha-mannosyltransferase |
| M. gilvum PYR-GCK | Mflv_2961 | - | 2e-21 | 28.49% (365) | glycosyl transferase, group 1 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2211c | - | 1e-22 | 30.37% (326) | hypothetical protein Mb2211c |
| M. tuberculosis H37Rv | Rv2188c | - | 1e-22 | 30.37% (326) | hypothetical protein Rv2188c |
| M. leprae Br4923 | MLBr_00886 | - | 4e-23 | 28.53% (375) | putative glycosyl transferase |
| M. abscessus ATCC 19977 | MAB_4057c | - | 2e-20 | 29.28% (321) | glycosyl transferase |
| M. marinum M | MMAR_3232 | - | 9e-22 | 27.47% (375) | glycosyltransferase |
| M. avium 104 | MAV_2306 | - | 1e-22 | 30.53% (285) | glycosyl transferase, group 1 |
| M. smegmatis MC2 155 | MSMEG_4253 | - | 1e-20 | 29.87% (318) | glycosyl transferase, group 1 family protein |
| M. thermoresistible (build 8) | TH_3188 | - | 2e-20 | 32.76% (293) | PUTATIVE glycosyl transferase, group 1 |
| M. ulcerans Agy99 | MUL_3543 | - | 2e-21 | 27.73% (375) | glycosyltransferase |
| M. vanbaalenii PYR-1 | Mvan_5246 | - | 0.0 | 87.66% (381) | glycosyl transferase, group 1 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1500|M.gilvum_PYR-GCK --------------------MTGGVSALVLIDAF--RMGGAETLLAPMIV
Mvan_5246|M.vanbaalenii_PYR-1 --------------------MTRGISALVLIDAF--RMGGAETLLAPMIV
Mb2211c|M.bovis_AF2122/97 --------------------MSR---VLLVTNDFPPRRGGIQSYLGEFVG
Rv2188c|M.tuberculosis_H37Rv --------------------MSR---VLLVTNDFPPRRGGIQSYLGEFVG
MMAR_3232|M.marinum_M --------------------MSR---VLLVTNDFPPRRGGIQSYLSEFVG
MUL_3543|M.ulcerans_Agy99 --------------------MSR---VLLVTNDFPPRRGGIQSYLSESVG
MLBr_00886|M.leprae_Br4923 --------------------MSR---VLLVTNDFPPRRGGIQSYLGEFVD
MAV_2306|M.avium_104 --------------------MSR---VLLVTNDFPPRPGGIHSYLGEFVG
MSMEG_4253|M.smegmatis_MC2_155 --------------------------MLLVTNDFPPRRGGIQSYLEAFVG
MAB_4057c|M.abscessus_ATCC_199 MLSGDEFARRAVAALKPRRIAVLSVHTSPLAQPGTGDAGGMNVYVLQTAL
TH_3188|M.thermoresistible__bu ---------------------MVSEHASPLAALGGVDAGGQNVHVAELSA
: ** . :
Mflv_1500|M.gilvum_PYR-GCK ASRDTDVSMDVVSISPSEWNSEKTMTILAEAGIPTRSLGIRRLLDPVALP
Mvan_5246|M.vanbaalenii_PYR-1 ASRDTDVTMDVVSISPADWNSEKTMAILADAGIATRSLGIRRLLDPVALP
Mb2211c|M.bovis_AF2122/97 RLVGSRAHAMTVYAPQWKGADAFDDAARAAGYRVVRHPSTVMLPGPTVDV
Rv2188c|M.tuberculosis_H37Rv RLVGSRAHAMTVYAPQWKGADAFDDAARAAGYRVVRHPSTVMLPGPTVDV
MMAR_3232|M.marinum_M RLVEAGSHSITVYAPQWKGAQGYDDVARAAGYRVVRHPSTLMLPVPTVDS
MUL_3543|M.ulcerans_Agy99 RLVEAGSHSITVYAPQWKGAQGYDDVARAAGYRVVRHPSTLMLPVPTVDS
MLBr_00886|M.leprae_Br4923 RLINIGSHDVMVYAPQWTGADTFDNAAHDGGYQVVRHPGTLMLPGPAVDA
MAV_2306|M.avium_104 RLAAAGSHELTVYAPDWKGAQAHDEAV---GYRVVRHPGTLMLPGPAVDA
MSMEG_4253|M.smegmatis_MC2_155 ELVRT--HELTVYAPKWKGAEEYDEKAARSGYRVVRHPTTLMLPEPTVAS
MAB_4057c|M.abscessus_ATCC_199 QLARRGVAVDIFTRATSSSDEPVVSVTDGVTVRNIVAGPFEGLDKYDLPT
TH_3188|M.thermoresistible__bu ALTRRGHEVTVYTRRDDRGLPDRVDTPLGYTVVHVPAGPAEQLPKDELLP
*
Mflv_1500|M.gilvum_PYR-GCK RLV------------------RTIRQGRYDVVHAHLEMAMTLAVPAAALA
Mvan_5246|M.vanbaalenii_PYR-1 RLV------------------KTIRQGRYDIVHAHLEMAMTLAVPAAALV
Mb2211c|M.bovis_AF2122/97 RMR------------------RLIAEHDIETVWFGAAAPLALLAPRARLA
Rv2188c|M.tuberculosis_H37Rv RMR------------------RLIAEHDIETVWFGAAAPLALLAPRARLA
MMAR_3232|M.marinum_M RMR------------------RLIAQHDIDTVWFGAAAPLALLAPRARQA
MUL_3543|M.ulcerans_Agy99 RMR------------------RLIAQHDIDTVWFGAAAPLALLAPRARQA
MLBr_00886|M.leprae_Br4923 RMR------------------RLIVDHGIDTVWFGSAAPLALLAPCARKA
MAV_2306|M.avium_104 RMR------------------RLIAEHVIDTVWFGAAAPLALLAQRARAA
MSMEG_4253|M.smegmatis_MC2_155 RMK------------------RLIGEHDIETVWFGAAAPLALLGPLARRA
MAB_4057c|M.abscessus_ATCC_199 QLCAFTAGVLRAEAVHEPGYYNLVHSHYWLSGQAGWLARDRWGVPLVHTA
TH_3188|M.thermoresistible__bu YMGPFAQFLDAEWSAHRP---DVVHAHFWMSGIATQLASRHLDLPAVQTF
: : .
Mflv_1500|M.gilvum_PYR-GCK GRPAVCTFHHVARPLEGRAAWRERLAVEAATRSRRALFVSEASRLSFAQN
Mvan_5246|M.vanbaalenii_PYR-1 GRPAVCTFHHVARPLQGRAAWRERLAVEAATRSRRALFVSEASRRSFAET
Mb2211c|M.bovis_AF2122/97 GASRVLASTHGHEVGWSMLPVARSVLRRIGDGTDVVTFVSSYTRSRFASA
Rv2188c|M.tuberculosis_H37Rv GASRVLASTHGHEVGWSMLPVARSVLRRIGDGTDVVTFVSSYTRSRFASA
MMAR_3232|M.marinum_M GATRVLASTHGHEVGWSMLPVARSVLRRIGDTTDIVTFVSRYTRSRFAPA
MUL_3543|M.ulcerans_Agy99 GATRVLASTHGHEVGWSMLPVAGSVLRRIGDTTDIVTFVSRYTRSRFAPA
MLBr_00886|M.leprae_Br4923 GATRVLASTHGHEVGWSMLPVARSVLRRIGDVTDVVTFVSRYTRSRVTAA
MAV_2306|M.avium_104 GARRVLASTHGHEVGWSMLPVARSVLRRIGEQTDVVTFVSRYTRSRFAPA
MSMEG_4253|M.smegmatis_MC2_155 GARRIVASTHGHEVGWSMLPVARTALRRIGNDADVVTFVSRYTRSRFASA
MAB_4057c|M.abscessus_ATCC_199 HTLAAVKNQTLAEGDRPEPALRGVGEQQVVDEADRLIVNTKDEADQLVSI
TH_3188|M.thermoresistible__bu HALGVVKKRHQGAEDTS-PQERLQLEAMVARGANWVAATCTDEVFELMRM
: .
Mflv_1500|M.gilvum_PYR-GCK YRPKGMPANWEVMHNGIDIANFAPGQADPGVRSELAGRD--GDSLVAVLP
Mvan_5246|M.vanbaalenii_PYR-1 YRPNRMPKNWEVMHNGIDISNFSPGQADPRVRSELAAG---TQGPVVVLP
Mb2211c|M.bovis_AF2122/97 FGP---AASLEYLPPGVDTDRFRP---DPAARAELRKRYRLGERPTVVCL
Rv2188c|M.tuberculosis_H37Rv FGP---AASLEYLPPGVDTDRFRP---DPAARAELRKRYRLGERPTVVCL
MMAR_3232|M.marinum_M FGP---AASLEYLPPGVDTDRFRP---DPAARAELRKRYQLGERPTVVCV
MUL_3543|M.ulcerans_Agy99 FGP---AASLEYLPPGVDTDRFRP---DPAARAELRKRYQLGERPTVVCV
MLBr_00886|M.leprae_Br4923 FGP---RAALEYLPPGVDADRFRP---DPASRAELRNRYRLGERPTVVCL
MAV_2306|M.avium_104 FGP---RARLEYLPSGVDTDRFRP---DPAGRARLRERYGLGERPTVVCV
MSMEG_4253|M.smegmatis_MC2_155 FGP---SAALEHLPPGVDTDRFAP---DPDARARMRERYGLGDRPVVVCL
MAB_4057c|M.abscessus_ATCC_199 HNADP--ARIDIVHPGVDLDVFTP----GDKAAARAEFGLRADEQVVAFV
TH_3188|M.thermoresistible__bu GRSR---SRISVVPCGVDLDLFTP----DGPQAPRGER------RRIVTV
. . : *:* * * : .
Mflv_1500|M.gilvum_PYR-GCK AAFRDFKGIPVAIKAWPAVIEKFPSAVLALTGGGE--------LESELRQ
Mvan_5246|M.vanbaalenii_PYR-1 AAFRDFKGIPVAIKAWPAVKQRFPTAVLALTGGGE--------LEGELRR
Mb2211c|M.bovis_AF2122/97 SRLVPRKGQDTLVTALPSIRRRVDGAALVIVGGGP--------YLETLRK
Rv2188c|M.tuberculosis_H37Rv SRLVPRKGQDTLVTALPSIRRRVDGAALVIVGGGP--------YLETLRK
MMAR_3232|M.marinum_M SRLVPRKGQDTLIKALPAIRRRVDGAALVIVGGGP--------YLETLRK
MUL_3543|M.ulcerans_Agy99 SRLVPRKGQDTLIKALPAIRRRVDGAALVIVGGGP--------YLETLRK
MLBr_00886|M.leprae_Br4923 ARLVPRKGQDMLIRALPSIRQRADGAALMIVGGGP--------YLGTLRK
MAV_2306|M.avium_104 SRLVPRKGQDMLIRAMPSIVQRVDGAALVIVGGGP--------HLQSLRA
MSMEG_4253|M.smegmatis_MC2_155 SRLVPRKGQDMLIRALPELRRRVPDTALAIVGGGP--------YLETLQR
MAB_4057c|M.abscessus_ATCC_199 GRIQPLKAPDLLVRAAERLPG----VRVLIVGGPSGSGLD---EPTALQD
TH_3188|M.thermoresistible__bu GRFVPRKGFDTLVRALPAIPN----AELVVVGGPRRSQLRTEPEARRLCT
. : *. : * : . : :.** *
Mflv_1500|M.gilvum_PYR-GCK LVADLGLGDSVNFVG--VRTDMPEVYRAADVVVLPS-THG-----ENLPT
Mvan_5246|M.vanbaalenii_PYR-1 LVADLGLGDSVNFVG--VRTDMPEVYRAADVVVLPS-THG-----ENLPT
Mb2211c|M.bovis_AF2122/97 LAHDCGVADHVTFTGGVATDELPAHHALADVFAMPCRTRGAGMDVEGLGI
Rv2188c|M.tuberculosis_H37Rv LAHDCGVADHVTFTGGVATDELPAHHALADVFAMPCRTRGAGMDVEGLGI
MMAR_3232|M.marinum_M LARDCEVDEHVTFTGGVPGEELPTHHAMADVFAMPCRTRGSGMDVEGLGI
MUL_3543|M.ulcerans_Agy99 LARDCEVDEHVTFTSGVPGEELPTHHAMADVFAMPCRTRGSGMDVEGLGI
MLBr_00886|M.leprae_Br4923 LAQGCGVADHVIFAGGVAAVEVPAHYTLADVFAMPCRTRGGGMDVEGLGI
MAV_2306|M.avium_104 LARDCGVAAQVTFTGGIPAAELPTHHALADVFAMPCRTRGAGLDVEGLGI
MSMEG_4253|M.smegmatis_MC2_155 MASDLGVAEHVVFTRGIPAEELPAHHAMADVFAMPCRTRGAGLDVEGLGI
MAB_4057c|M.abscessus_ATCC_199 LAVDLGIADRVTFLPPQTRERLAQVYRAADIVAVPS-------YSESFGL
TH_3188|M.thermoresistible__bu LAEELGVADRVQLYGSVARADMPAVLRSADVVACTP-------WYEPFGI
:. : * : :. **:.. . * :
Mflv_1500|M.gilvum_PYR-GCK VLIESSATACAVAASRVGGIPDIVLDGQTGLLFEAGSVEGFADTVCRLLG
Mvan_5246|M.vanbaalenii_PYR-1 VLIEASATACAIAASRVGGIPDIVLDGATGLLFEPGSADGLAEAVCRLLD
Mb2211c|M.bovis_AF2122/97 VFLEASAAGVPVIAGNSGGAPETVQHNKTGLVVDGRSVDRVADAVAELLI
Rv2188c|M.tuberculosis_H37Rv VFLEASAAGVPVIAGNSGGAPETVQHNKTGLVVDGRSVDRVADAVAELLI
MMAR_3232|M.marinum_M VFLEASASGVPVIAGESGGAPETVQHNKTGLVVDGNSVDKVAGAVIEVLA
MUL_3543|M.ulcerans_Agy99 VFLEASASGVPVIAGESGGAPETVQHNKTGLVVDGNSVDKVAGAVIEVLA
MLBr_00886|M.leprae_Br4923 VFLEASATGVPVIAGKSGGAPEAVQHNKTGLVVDGRSVNMVADAVTELLT
MAV_2306|M.avium_104 VYLEASASGVPVIAGRSGGAPETVQHNKTGLVVDGRSVTEIADAVTELLS
MSMEG_4253|M.smegmatis_MC2_155 VYLEASACGVPVVAGRSGGAPETVLDGKTGTVVDGTDVDAITTAVGDLLA
MAB_4057c|M.abscessus_ATCC_199 VAIEAQACGTPVVAAAVGGLPVAVADQRTGLLVPTHRTEDWADAIGELLV
TH_3188|M.thermoresistible__bu VPLEAMACGVPVVAAAVGGMLDTVVHDVTGRLVPPRRPAELADTINQLLR
* :*: * . .: *. ** * . ** :. : :: :*
Mflv_1500|M.gilvum_PYR-GCK DAELRRRLGAAASERARSEFSATAWLARLRTTYLEAMGDRR---------
Mvan_5246|M.vanbaalenii_PYR-1 DVELRRRMGAAASERARTEFSATAWLARLRTTYLEAMGVPR---------
Mb2211c|M.bovis_AF2122/97 DRDRAVAMGAAGREWVTAQWRWDTLAAKLADFLRGDDAAR----------
Rv2188c|M.tuberculosis_H37Rv DRDRAVAMGAAGREWVTAQWRWDTLAAKLADFLRGDDAAR----------
MMAR_3232|M.marinum_M DRDRAARMGDAGRQWVTSQWRWDTLAARLAELLRGDDTGR----------
MUL_3543|M.ulcerans_Agy99 DRDRAARMGDAGRQWVTSQWRWDTLAARLAELLRGDDAGR----------
MLBr_00886|M.leprae_Br4923 DRDRAAAMGAAGRHWVTAEWRWDTLAARLADLLCGNDSR-----------
MAV_2306|M.avium_104 DPDRAAAMGAAGRQWVTSQWRWDTLAARLAGLLGD---------------
MSMEG_4253|M.smegmatis_MC2_155 DPRRAAAMGVAGRHWALDNWQWRTRGARLAELLSGRREARQA--------
MAB_4057c|M.abscessus_ATCC_199 RKGAGFSRAAVEHAAG---FSWSSTADSLLSSYGRAIADYRAPQRPSTQW
TH_3188|M.thermoresistible__bu DSFLRQSLGAAGRDRARARYSWDRVATDTQRIYDRLVPADYRSATASQMS
. . :
Mflv_1500|M.gilvum_PYR-GCK ----------------
Mvan_5246|M.vanbaalenii_PYR-1 ----------------
Mb2211c|M.bovis_AF2122/97 ----------------
Rv2188c|M.tuberculosis_H37Rv ----------------
MMAR_3232|M.marinum_M ----------------
MUL_3543|M.ulcerans_Agy99 ----------------
MLBr_00886|M.leprae_Br4923 ----------------
MAV_2306|M.avium_104 ----------------
MSMEG_4253|M.smegmatis_MC2_155 ----------------
MAB_4057c|M.abscessus_ATCC_199 ASRARFRPRRLSGLRR
TH_3188|M.thermoresistible__bu SG--------------