For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
STSRVAIVHERFTEFGGSELVVAEFMKTWPQAKVFAPITEPHCRRQVMHAAGRTEGAPHEPISGTWLDDA YALTGRRSHAPLLPLVPGALGKLPLGDDVDAVLISHHSFATQAAFATEAPVVAYVHSPARWAWDRTFRDH EMASRAGRIALSALGKLARRGELRAAPRLSHVIANSHAVAERIRDWWGLPSTVISPPVRIDRFSPDPSIR REDFYLCAGRLVPYKRADLAIRAAQRANCRLVVLGEGRFREHLEDIAGPETTFLGAASDDVLLDMYRRCR ALLMPGVEDFGIVPVEAMACGTPVLALGEGGALDTVTPGITGEHMRAGPDDAVVEQLAALMRDFDPADYD AGTIRARACEFSPEAFRARIAETVQGAVRA*
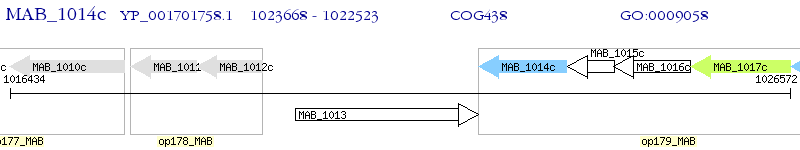
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1014c | - | - | 100% (381) | glycosyl transferase |
| M. abscessus ATCC 19977 | MAB_1976 | - | 4e-11 | 29.86% (211) | hypothetical protein MAB_1976 |
| M. abscessus ATCC 19977 | MAB_4057c | - | 6e-11 | 33.77% (151) | glycosyl transferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2211c | - | 1e-12 | 32.42% (182) | hypothetical protein Mb2211c |
| M. gilvum PYR-GCK | Mflv_0483 | - | 1e-10 | 29.80% (198) | glycosyl transferase, group 1 |
| M. tuberculosis H37Rv | Rv2188c | - | 2e-12 | 32.42% (182) | hypothetical protein Rv2188c |
| M. leprae Br4923 | MLBr_00886 | - | 7e-10 | 30.77% (182) | putative glycosyl transferase |
| M. marinum M | MMAR_3232 | - | 9e-12 | 30.52% (213) | glycosyltransferase |
| M. avium 104 | MAV_4082 | - | 3e-15 | 33.21% (271) | transferase |
| M. smegmatis MC2 155 | MSMEG_5641 | - | 2e-12 | 29.55% (247) | glycosyl transferase, group 1 family protein |
| M. thermoresistible (build 8) | TH_0092 | - | 6e-17 | 30.93% (291) | glycosyl transferase, group 1 family protein |
| M. ulcerans Agy99 | MUL_3543 | - | 3e-11 | 30.05% (213) | glycosyltransferase |
| M. vanbaalenii PYR-1 | Mvan_0978 | - | 7e-11 | 31.98% (197) | glycosyl transferase, group 1 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_3232|M.marinum_M MSRVLLVTN-------------DFPPRRGGIQSYLSEFVGRLVEAGSHSI
MUL_3543|M.ulcerans_Agy99 MSRVLLVTN-------------DFPPRRGGIQSYLSESVGRLVEAGSHSI
Mb2211c|M.bovis_AF2122/97 MSRVLLVTN-------------DFPPRRGGIQSYLGEFVGRLVGSRAHAM
Rv2188c|M.tuberculosis_H37Rv MSRVLLVTN-------------DFPPRRGGIQSYLGEFVGRLVGSRAHAM
MLBr_00886|M.leprae_Br4923 MSRVLLVTN-------------DFPPRRGGIQSYLGEFVDRLINIGSHDV
MSMEG_5641|M.smegmatis_MC2_155 MSGN------------------------DILAPADVDDPLRIVLVAPPYF
TH_0092|M.thermoresistible__bu MSGNGFRFHHTFRDQNATAADTAPADTAALLSETESTDPLRIVLVAPPYF
Mflv_0483|M.gilvum_PYR-GCK ---------------------------------------MRVALIAPHHF
MAV_4082|M.avium_104 MRIAMISEHASPLAHLGGVDAGGQNVHVAELSCALARRGHDVVVYTRRDD
Mvan_0978|M.vanbaalenii_PYR-1 MRVAIVAES--------------FLPNVNGVTNSVLRVIEHLRRTGHEVL
MAB_1014c|M.abscessus_ATCC_199 MSTSRVAIVHERFTEFGG----SELVVAEFMKTWPQAKVFAPITEPHCRR
MMAR_3232|M.marinum_M TVYAPQWKGAQGYDDVARAAGYRVVR---HPSTLMLPVPTVDSRMRRLIA
MUL_3543|M.ulcerans_Agy99 TVYAPQWKGAQGYDDVARAAGYRVVR---HPSTLMLPVPTVDSRMRRLIA
Mb2211c|M.bovis_AF2122/97 TVYAPQWKGADAFDDAARAAGYRVVR---HPSTVMLPGPTVDVRMRRLIA
Rv2188c|M.tuberculosis_H37Rv TVYAPQWKGADAFDDAARAAGYRVVR---HPSTVMLPGPTVDVRMRRLIA
MLBr_00886|M.leprae_Br4923 MVYAPQWTGADTFDNAAHDGGYQVVR---HPGTLMLPGPAVDARMRRLIV
MSMEG_5641|M.smegmatis_MC2_155 DIPPKGYGGTEAVVADLADTLTARGH---HVTVLGAGAPGTAARFQPLWE
TH_0092|M.thermoresistible__bu DIPPKAYGGTEAVVADLADSLTARGH---RVTVLGAGNPRIAARFVPLWE
Mflv_0483|M.gilvum_PYR-GCK PIREPFAGGIESHVWHLARALIRQGH---AVTLCAAAGSDLAMEHPALTV
MAV_4082|M.avium_104 PRLPDSVETPHGYSVVHVPAGPARVL---PKDELLQYMYGFAQFLRQQWC
Mvan_0978|M.vanbaalenii_PYR-1 VIAPDTPRGQPAADKVYEGVRVHRVPSMMFPKITSLPLGVPRPRMVGVLR
MAB_1014c|M.abscessus_ATCC_199 QVMHAAGRTEGAPHEPISGTWLDDAYALTGRRSHAPLLPLVPGALGKLPL
MMAR_3232|M.marinum_M QHDIDTVWFG-------AAAPLALLAPRARQAGATRVLASTHGHEVGWS-
MUL_3543|M.ulcerans_Agy99 QHDIDTVWFG-------AAAPLALLAPRARQAGATRVLASTHGHEVGWS-
Mb2211c|M.bovis_AF2122/97 EHDIETVWFG-------AAAPLALLAPRARLAGASRVLASTHGHEVGWS-
Rv2188c|M.tuberculosis_H37Rv EHDIETVWFG-------AAAPLALLAPRARLAGASRVLASTHGHEVGWS-
MLBr_00886|M.leprae_Br4923 DHGIDTVWFG-------SAAPLALLAPCARKAGATRVLASTHGHEVGWS-
MSMEG_5641|M.smegmatis_MC2_155 RTLPDRLGQP-------YPEIMHALKVRKAIERIAATEG-VDIVHDHTF-
TH_0092|M.thermoresistible__bu RTIPDRLGQP-------YPEVMHALKVRRAIADIADREGGIDLIHDHTF-
Mflv_0483|M.gilvum_PYR-GCK TAMPRSAAASRVFPPPGAIKESAHHAFVELMSELADPASGVDVVHNHSLN
MAV_4082|M.avium_104 SDGPDVAHAHFWMSG--VATQQAARAHSIPTVQTFHALGLTKRLHQGSDD
Mvan_0978|M.vanbaalenii_PYR-1 GFDPDVVHLASPALLG-WGGVHAARRLGIPTVAVFQTDVAGFAESYGIG-
MAB_1014c|M.abscessus_ATCC_199 GDDVDAVLISHHS----FATQAAFATEAPVVAYVHSPARWAWDRTFRDHE
MMAR_3232|M.marinum_M MLPVA-RSVLRRIGDTTDIVTFVSRYTRSRFAPAFGPAASLEYLPPGVDT
MUL_3543|M.ulcerans_Agy99 MLPVA-GSVLRRIGDTTDIVTFVSRYTRSRFAPAFGPAASLEYLPPGVDT
Mb2211c|M.bovis_AF2122/97 MLPVA-RSVLRRIGDGTDVVTFVSSYTRSRFASAFGPAASLEYLPPGVDT
Rv2188c|M.tuberculosis_H37Rv MLPVA-RSVLRRIGDGTDVVTFVSSYTRSRFASAFGPAASLEYLPPGVDT
MLBr_00886|M.leprae_Br4923 MLPVA-RSVLRRIGDVTDVVTFVSRYTRSRVTAAFGPRAALEYLPPGVDA
MSMEG_5641|M.smegmatis_MC2_155 AGPLN-APFYRKLG-LPTVVTVHGPIDEDLYPYYRELGDDIGLIAISDRQ
TH_0092|M.thermoresistible__bu AGPLN-VPAYRELG-VPTVATVHGPIDEDLYPYYRELGHDVGLIAISDRQ
Mflv_0483|M.gilvum_PYR-GCK PVPVL-VARRLPVPVLTTLHTPPFPALEAAVGAVARSGQAFAAVSRQAAA
MAV_4082|M.avium_104 TSPDCRIDVEARVAREADWVAATSTDEVFELVRMGRARSRTSVVPCGVDL
Mvan_0978|M.vanbaalenii_PYR-1 MLSHASWAWTRRLHSKADRTLAPSTSAMENLAAHRIPR--VHKWARGVDV
MAB_1014c|M.abscessus_ATCC_199 MASRAGRIALSALGKLARRGELRAAPRLSHVIANSHAVAERIRDWWGLPS
. : .
MMAR_3232|M.marinum_M DRFRPDPAARAELRKRYQLGERPTVVCVSRLVPRKGQDTLIKALP----A
MUL_3543|M.ulcerans_Agy99 DRFRPDPAARAELRKRYQLGERPTVVCVSRLVPRKGQDTLIKALP----A
Mb2211c|M.bovis_AF2122/97 DRFRPDPAARAELRKRYRLGERPTVVCLSRLVPRKGQDTLVTALP----S
Rv2188c|M.tuberculosis_H37Rv DRFRPDPAARAELRKRYRLGERPTVVCLSRLVPRKGQDTLVTALP----S
MLBr_00886|M.leprae_Br4923 DRFRPDPASRAELRNRYRLGERPTVVCLARLVPRKGQDMLIRALP----S
MSMEG_5641|M.smegmatis_MC2_155 RELAPDLNWVGRVHNALRIDDWPFNTVKKDYALFLGRYAPYKGAH----L
TH_0092|M.thermoresistible__bu RELAPDLNWIGRVHNALRIHDWPFKREKGDYALFLGRFAPYKGAH----L
Mflv_0483|M.gilvum_PYR-GCK AWEHVGIRRMALVRNGIDTSRWPAGPG-GEDLVWSGRLLEEKGPH----L
MAV_4082|M.avium_104 DAFTPDGPVAAR-------GARPRIVTVGRLVPRKGFDTIVRALPRIPDA
Mvan_0978|M.vanbaalenii_PYR-1 TGFAPS-ARDERLRRSWSPDGRPIIGFVGRLAPEKHVERL---------A
MAB_1014c|M.abscessus_ATCC_199 TVISPP-VRIDRFSPDPSIRREDFYLCAGRLVPYKRADLAIRAAQ-----
MMAR_3232|M.marinum_M IRRRVDGAALVIVGGGPYLETLRKLARDCEVDEHVTFTGGVPGEELPTHH
MUL_3543|M.ulcerans_Agy99 IRRRVDGAALVIVGGGPYLETLRKLARDCEVDEHVTFTSGVPGEELPTHH
Mb2211c|M.bovis_AF2122/97 IRRRVDGAALVIVGGGPYLETLRKLAHDCGVADHVTFTGGVATDELPAHH
Rv2188c|M.tuberculosis_H37Rv IRRRVDGAALVIVGGGPYLETLRKLAHDCGVADHVTFTGGVATDELPAHH
MLBr_00886|M.leprae_Br4923 IRQRADGAALMIVGGGPYLGTLRKLAQGCGVADHVIFAGGVAAVEVPAHY
MSMEG_5641|M.smegmatis_MC2_155 ALEAGHAAGMPLVLAGKCSEPPEIAYFDEQVKPLLTDSDHVFG--MADAQ
TH_0092|M.thermoresistible__bu AIQAADRAGIRLVLAGKCDEPSEKAYFEEQVRPLLNENDEVFG--EADAI
Mflv_0483|M.gilvum_PYR-GCK AVDAALRAGRRIVLAGPIVDD---HFFRRRIAPTLGSRASYAG--HLGQR
MAV_4082|M.avium_104 ELVIVGGPPAAELAGDEQARRLQRLAGELGVADRVRLLGGVTRAQMPALL
Mvan_0978|M.vanbaalenii_PYR-1 ALHARDDLQIVVVGDG------VDRAKLESELPRAVFTGALYGDELAAAY
MAB_1014c|M.abscessus_ATCC_199 ----RANCRLVVLGEGR-----FREHLEDIAGPETTFLGAASDDVLLDMY
: .
MMAR_3232|M.marinum_M AMADVFAMPCRTRGSGMDVEGLGIVFLEASASGVPVIAGESGGAPETVQH
MUL_3543|M.ulcerans_Agy99 AMADVFAMPCRTRGSGMDVEGLGIVFLEASASGVPVIAGESGGAPETVQH
Mb2211c|M.bovis_AF2122/97 ALADVFAMPCRTRGAGMDVEGLGIVFLEASAAGVPVIAGNSGGAPETVQH
Rv2188c|M.tuberculosis_H37Rv ALADVFAMPCRTRGAGMDVEGLGIVFLEASAAGVPVIAGNSGGAPETVQH
MLBr_00886|M.leprae_Br4923 TLADVFAMPCRTRGGGMDVEGLGIVFLEASATGVPVIAGKSGGAPEAVQH
MSMEG_5641|M.smegmatis_MC2_155 AKRKLLAEARCLVFPIQWEEPFGMVMIEAMACGTPVVALRGGAVPEVIVD
TH_0092|M.thermoresistible__bu AKRELLANARCLLFPIQWEEPFGMVMIEAMACGTPVVALRGGAVPEVIVD
Mflv_0483|M.gilvum_PYR-GCK ELATLVGRAAAALVTPLWDEPYGQVVVEAMACGTPVVAFAGGGIPELVDE
MAV_4082|M.avium_104 RSADVVACTP-------WYEPFGIVPLEAMACGVPVVATAVGGIRDTVVD
Mvan_0978|M.vanbaalenii_PYR-1 ASMDVFVHPG-------EHETFCQAVQEAMASGLPVIAPDAGGPRDLVAP
MAB_1014c|M.abscessus_ATCC_199 RRCRALLMPG--------VEDFGIVPVEAMACGTPVLALGEGGALDTVTP
. . * . ** * * **:* *. : :
MMAR_3232|M.marinum_M NKTGLVVDG----------NSVDKVAGAVIEVLADRDRAARMGDAGRQWV
MUL_3543|M.ulcerans_Agy99 NKTGLVVDG----------NSVDKVAGAVIEVLADRDRAARMGDAGRQWV
Mb2211c|M.bovis_AF2122/97 NKTGLVVDG----------RSVDRVADAVAELLIDRDRAVAMGAAGREWV
Rv2188c|M.tuberculosis_H37Rv NKTGLVVDG----------RSVDRVADAVAELLIDRDRAVAMGAAGREWV
MLBr_00886|M.leprae_Br4923 NKTGLVVDG----------RSVNMVADAVTELLTDRDRAAAMGAAGRHWV
MSMEG_5641|M.smegmatis_MC2_155 GVTGVICDVPEELPAAME-KATHLDPAACRRHVAAHFGVGRFGAGYEQIY
TH_0092|M.thermoresistible__bu GVTGVICEDPAELPAAIE-RTADMDPAECRRHVAANFDIERFGSGYEQIF
Mflv_0483|M.gilvum_PYR-GCK HSGRLVEPGDVAALAAAVREAVALPRAGVRRVAVQRHGLARMVTGYVALY
MAV_4082|M.avium_104 GATGRLVPP----------KDPDALGEAVAALLRDGRRGRTLGEAGRERA
Mvan_0978|M.vanbaalenii_PYR-1 YRTGLLLPVA---------EFESRLAESVDHLVAERRR--YSVAARRSVL
MAB_1014c|M.abscessus_ATCC_199 GITGEHMRAG----------PDDAVVEQLAALMRDFDPADYDAGTIRARA
MMAR_3232|M.marinum_M TSQWR-------WDTLAARLAELLRGDDTGR----------------
MUL_3543|M.ulcerans_Agy99 TSQWR-------WDTLAARLAELLRGDDAGR----------------
Mb2211c|M.bovis_AF2122/97 TAQWR-------WDTLAAKLADFLRGDDAAR----------------
Rv2188c|M.tuberculosis_H37Rv TAQWR-------WDTLAAKLADFLRGDDAAR----------------
MLBr_00886|M.leprae_Br4923 TAEWR-------WDTLAARLADLLCGNDSR-----------------
MSMEG_5641|M.smegmatis_MC2_155 RNVLR----QRTRMTLREVLTEPPGVAERASA---------------
TH_0092|M.thermoresistible__bu RGVVDGSMAVRSNVALRQILPETGERDEQIA----------------
Mflv_0483|M.gilvum_PYR-GCK RALIAEATPPPRPPKLHSSSESANQSQAVSATARACLTNVSGVSAET
MAV_4082|M.avium_104 RARYSWDRVAADTERIYERLSPARRGVGTPSTRSG------------
Mvan_0978|M.vanbaalenii_PYR-1 GRTWP-----VICDELLGHYEAVMGARRLRAA---------------
MAB_1014c|M.abscessus_ATCC_199 CEFSP--------EAFRARIAETVQGAVRA-----------------
: