For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
INAIANGLVGSVRAAHRRKMLLSGIALAAVLLIAIGYLTVGALRVSPFSGTYRVKVQLPESGGLLPNQDV ALRGVKIGTVQSLDITQQGVDAVAEIQSKYKIPVSAKVRVTGLSPAGEQYINFEGVSDQGPFLKDGAVIS QGAQVPVTLAKLLADADGLLAQVDPKKLELIKKELSLSKEGPRKLTEIIDGGTFLLNTLDSVLPETTSIL RSSRVVLTLASDKNSGIQATANELGRTLRGIDKMQNGYRTLVDRTPNTLHSVDNLFDDNSDTMVTLLGSL ATASKLLYLRVPALNALFPNYRGSVFDALMSIMHEGGIWATADLYPRYVCDYPTPRLPGSAADYPEPFLY ANYCADEDPAVLIRGAKNAPRPAGDDTAGPPMGADMGKKADPTPKGRFQVPTPYGGPTLPIEPPH*
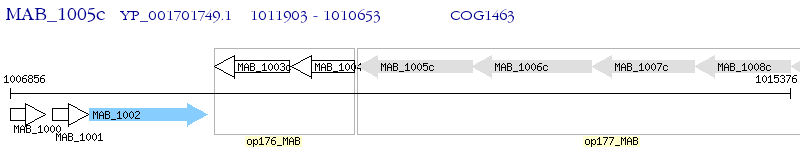
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1005c | - | - | 100% (416) | putative MCE family protein |
| M. abscessus ATCC 19977 | MAB_4511c | - | 0.0 | 75.72% (416) | putative Mce family protein |
| M. abscessus ATCC 19977 | MAB_4563c | - | e-150 | 61.54% (416) | putative Mce family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0180 | mce1F | 5e-25 | 25.13% (378) | MCE-family protein MCE1F |
| M. gilvum PYR-GCK | Mflv_0005 | - | 1e-157 | 64.42% (416) | virulence factor Mce family protein |
| M. tuberculosis H37Rv | Rv0174 | mce1F | 5e-25 | 25.13% (378) | MCE-family protein MCE1F |
| M. leprae Br4923 | MLBr_02594 | mce1F | 4e-20 | 21.66% (374) | putative secreted protein |
| M. marinum M | MMAR_3869 | mce5F | 1e-162 | 66.91% (417) | Mce family protein Mce5F |
| M. avium 104 | MAV_2383 | - | 1e-164 | 67.86% (420) | mce related protein |
| M. smegmatis MC2 155 | MSMEG_1148 | - | 1e-151 | 63.40% (418) | mce related protein |
| M. thermoresistible (build 8) | TH_2636 | - | 1e-107 | 60.54% (332) | PUTATIVE mce related protein |
| M. ulcerans Agy99 | MUL_3801 | mce5F | 1e-161 | 66.43% (417) | Mce family protein Mce5F |
| M. vanbaalenii PYR-1 | Mvan_0964 | - | 1e-154 | 63.94% (416) | hypothetical protein Mvan_0964 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_3869|M.marinum_M MITPIA---NAVVRVVRAGHRQQTWLSVAGLVLTLVVAAAYLLVGALRLT
MUL_3801|M.ulcerans_Agy99 MITPIA---NAVVRVVRAGHRQQTWLSVAGLVLTLVVAAAYLLVGALRLT
MAV_2383|M.avium_104 MIRAVAGFANLVVRAVRTGHRQQVWLSVAGLVLILVVATAYLLIGALRVT
MAB_1005c|M.abscessus_ATCC_199 MINAIA---NGLVGSVRAAHRRKMLLSGIALAAVLLIAIGYLTVGALRVS
Mflv_0005|M.gilvum_PYR-GCK MINALA---DSVVRVVRTVHRGRAWLSAGALVFILVVAAAYLFVGALQVN
Mvan_0964|M.vanbaalenii_PYR-1 MIGSVA---DAVVGVARFGYRRRAWLSTAALIMTLVVASAYLFFGALQVN
MSMEG_1148|M.smegmatis_MC2_155 MINAVA---DRIVGLVRFGYRKRSWLSALGLVMTLVVATAYLFFGALQVN
TH_2636|M.thermoresistible__bu MIGAVA---DRIVALVRFGHRRRATLSTIGLILTLVVATAYLLFGALQVN
Mb0180|M.bovis_AF2122/97 -------------MLTRFIRRQLILFAIVSVVAIVVLGWYYLRIPSLVG-
Rv0174|M.tuberculosis_H37Rv -------------MLTRFIRRQLILFAIVSVVAIVVLGWYYLRIPSLVG-
MLBr_02594|M.leprae_Br4923 -------------MLTPLIRRQLVLFAILTVISLTVLGVYYLQIPSLVG-
. * :: : ::. ** . :*
MMAR_3869|M.marinum_M PFASSYRVTVSLPESGGLLPNQDVALRGVRIGRIESLHITESGVDAIASI
MUL_3801|M.ulcerans_Agy99 PFASSYRVTVSLPESGGLLPNQDVALRGVRIGRIESLHLTESGVDAIASI
MAV_2383|M.avium_104 PFASSYRVTVQLPESGGLLPNQDVALRGVRIGRIESLQITDNGVNAVAAI
MAB_1005c|M.abscessus_ATCC_199 PFSGTYRVKVQLPESGGLLPNQDVALRGVKIGTVQSLDITQQGVDAVAEI
Mflv_0005|M.gilvum_PYR-GCK PLAKSYRVTVQLPESAGLLPNQNVTLRGVPIGRVERLDITPAGVDAVVSM
Mvan_0964|M.vanbaalenii_PYR-1 PLASSYRVTVELPESAGLLPNQNVTMRGVPIGRVERLDITPAGVNAVVSV
MSMEG_1148|M.smegmatis_MC2_155 PLDSDYGLTVQLPESAGLLPDQDVTLRGVPIGRVERLDITPSGVNAIVKV
TH_2636|M.thermoresistible__bu PFASSYQVTVALPESGGLLPNQAVTLRGVPVGRVERLDITPEGVNAVVDI
Mb0180|M.bovis_AF2122/97 --IGQYTLKADLPASGGLYPTANVTYRGITIGKVTAVEPTDQGARVTMSI
Rv0174|M.tuberculosis_H37Rv --IGQYTLKADLPASGGLYPTANVTYRGITIGKVTAVEPTDQGARVTMSI
MLBr_02594|M.leprae_Br4923 --IGRYSLKAELSATGGLYPTANVTYRGIAIGKVTDVEPTATGAEATMSI
* :.. *. :.** * *: **: :* : :. * *. . :
MMAR_3869|M.marinum_M TSQVKIPASSAVHVSALSPAGEQFLDFE--PESDTGPYLGEGAVIGVDQA
MUL_3801|M.ulcerans_Agy99 TSQVKIPASSTVHVSALSPAGEQFLDFE--PESDTGPYLSEGAVIGVDQA
MAV_2383|M.avium_104 TSKVRIPANTVAHVSALSPAGEQFINFE--AASDAGPYLHDGSFITSERT
MAB_1005c|M.abscessus_ATCC_199 QSKYKIPVSAKVRVTGLSPAGEQYINFE--GVSDQGPFLKDGAVISQG-A
Mflv_0005|M.gilvum_PYR-GCK DSSVAVPMSSAVRVSGLSPAGEQYIDFV--ADSDAGPYLENGSVIAQDNT
Mvan_0964|M.vanbaalenii_PYR-1 DSSAAVPVASSVRVSGLSPAGEQYIDFV--ADSGDGPYLADGAVVSQDNT
MSMEG_1148|M.smegmatis_MC2_155 KSTVRIPEASDVRVSGLSPAGEQYIDFIDVPAEDSSRFLADGAVIRQGQA
TH_2636|M.thermoresistible__bu DSAVKIPDSSAVRVSGLSPAGEQYIDFI--PEEDTGRYLADGAHIGQGTA
Mb0180|M.bovis_AF2122/97 ASNYKIPVDASANVHSVSAVGEQYIDLVS--TGAPGKYFSSGQTITKG--
Rv0174|M.tuberculosis_H37Rv ASNYKIPVDASANVHSVSAVGEQYIDLVS--TGAPGKYFSSGQTITKG--
MLBr_02594|M.leprae_Br4923 DSRYKIPIDSAANVHSVSAVGEQYLDLVS--SGNPGKYLAPGQTITKS--
* :* : ..* .:*..***:::: . :: * :
MMAR_3869|M.marinum_M RVPVSLAQLLGDANGLLAQIDPHQVELIKKELSLSQAGP-KKLAAIVDGG
MUL_3801|M.ulcerans_Agy99 RVPVSLAQLLGDANGLRAQIDPHQVELIKKVLSLSQAGP-KKLAAIVDGG
MAV_2383|M.avium_104 TVPVSLAQLLGDADGLLAQVDPRKIELIKKELSLSKEGP-AKLTAIVDGG
MAB_1005c|M.abscessus_ATCC_199 QVPVTLAKLLADADGLLAQVDPKKLELIKKELSLSKEGP-RKLTEIIDGG
Mflv_0005|M.gilvum_PYR-GCK TVPVSLADLLANADGALSQVDPGKLEVIKHELSMTEAGP-QKLADIIDGG
Mvan_0964|M.vanbaalenii_PYR-1 VVPVSLADLLANADGALAQADPAKLELIKRELSMSEAGP-QKLADIIDGG
MSMEG_1148|M.smegmatis_MC2_155 TVPVSLADLLADADGALGQVDTAKIELIKRELSLTDAGP-QKLADIVDGG
TH_2636|M.thermoresistible__bu TVPVPLAELLADADGALAQADVEKLEIIRRELSLSPAGP-QKLKDIVDGG
Mb0180|M.bovis_AF2122/97 TVPSEIGPALDNSNRGLAALPTEKIGLLLDETAQAVGGLGPALQRLVDST
Rv0174|M.tuberculosis_H37Rv TVPSEIGPALDNSNRGLAALPTEKIGLLLDETAQAVGGLGPALQRLVDST
MLBr_02594|M.leprae_Br4923 KIPIEIGPALDSVNRGLAALPKEKIGSLLDETAQAVGGLGPALQRLVSST
:* :. * . : . :: : : : * * ::..
MMAR_3869|M.marinum_M TFLLSTLDSVLPETTSIIRTGRVVLNLVADKNAGLAATAVALDRTLAGVA
MUL_3801|M.ulcerans_Agy99 TFLLSTLDSVLPETTSIIRTSRVVLNLIADKNAGLAATAVTLDRTLAGVA
MAV_2383|M.avium_104 LFLLSTLDSVLPETTSIIKTSRVVLNLASDKNNGLGAAATELNHTLTGVA
MAB_1005c|M.abscessus_ATCC_199 TFLLNTLDSVLPETTSILRSSRVVLTLASDKNSGIQATANELGRTLRGID
Mflv_0005|M.gilvum_PYR-GCK TFLLTTLDSVLPETSSMLRTSRVVFTLAADKNAGIEVASDNLAQTFDGVN
Mvan_0964|M.vanbaalenii_PYR-1 TFLLTTLDSVLPETSSMLRTSRVVLTLAADKNAGIEVASQNLDQVFDGVN
MSMEG_1148|M.smegmatis_MC2_155 TFLLSTLDSVLPETSSLLRTSRVVFTLMADKNPGISVASDNLTETFTGMD
TH_2636|M.thermoresistible__bu TFLLSTLDSVLPETSSLLRTSRVVFTLIDDKNAGIDVAADNLADTFDGVN
Mb0180|M.bovis_AF2122/97 QAIVGDFKTNIGDVNDIIENSGPILDSQVNTGDQIERWARKLNNLAAQTA
Rv0174|M.tuberculosis_H37Rv QAIVGDFKTNIGDVNDIIENSGPILDSQVNTGDQIERWARKLNNLAAQTA
MLBr_02594|M.leprae_Br4923 QAIVDAFKTNITDVNDIIQNSGPILDSQVKSDNAIERWVHNLNVLAAQSA
:: :.: : :...::... :: ... : *
MMAR_3869|M.marinum_M RMQGGYRSLTERTPHTLAAIDHLFADNSDTMVQLLGSMATAAQLLYLRVP
MUL_3801|M.ulcerans_Agy99 RMQGGYRSLTERTPHTLAAIDHLFADNSDTMVQLLGSMATAAQLLYLRVP
MAV_2383|M.avium_104 RMQAGYRRLTSQTPQTLSAVDNLFADNSDTMVQLLGSMATMSQLLYLRVP
MAB_1005c|M.abscessus_ATCC_199 KMQNGYRTLVDRTPNTLHSVDNLFDDNSDTMVTLLGSLATASKLLYLRVP
Mflv_0005|M.gilvum_PYR-GCK RMRDGFRRLTDRAPGTLNSVDNLFADNSDTMVQLLGNLTTTSQLLYLRVP
Mvan_0964|M.vanbaalenii_PYR-1 RMRQGFRTLTEQTPETLAEVDNLFADNSETMVQLLGSLTSTSQLLYLRVP
MSMEG_1148|M.smegmatis_MC2_155 KMREGYRRLTNQTPETLAAVDNLFVDNSDTMVQLLGNLTTASRLLYLRVP
TH_2636|M.thermoresistible__bu RMREGYRRLTDQTPEVLAHVDNLFADNSDTMVQLLGNLTTTSRLLYLRVP
Mb0180|M.bovis_AF2122/97 TRDQNVRSILSQAAPTADEVNAVFSGVRDSLPQTLANLEVVFDMLKRYHA
Rv0174|M.tuberculosis_H37Rv TRDQNVRSILSQAAPTADEVNAVFSGVRDSLPQTLANLEVVFDMLKRYHA
MLBr_02594|M.leprae_Br4923 RQDQHLKSILSQAAPTADEVNTVFSDVRDSLPQTLANFEILIDMLKRYHH
: : .::. . :: :* . ::: *..: :*
MMAR_3869|M.marinum_M ALNALFPDYRG------SVLDALASAFHDHGVWATAELYPRYTCDYGTPS
MUL_3801|M.ulcerans_Agy99 ALNALFPDYRG------SVLDALASAFHDHGVWATAELYPRYTCDYGTPS
MAV_2383|M.avium_104 ALNALFPDYRG------SVLDAVTSVFHDHGVWATADLYPRYVCDYGTPA
MAB_1005c|M.abscessus_ATCC_199 ALNALFPNYRG------SVFDALMSIMHEGGIWATADLYPRYVCDYPTPR
Mflv_0005|M.gilvum_PYR-GCK ALHALFPDYRT------SVLDAIGSAMHDNGLWATGEIYPRYTCDYGTPR
Mvan_0964|M.vanbaalenii_PYR-1 ALNALFPAHRT------SVLDAIGSIMHDNGLWATGDIYPRYSCDYGTPR
MSMEG_1148|M.smegmatis_MC2_155 ALNALFPSYRT------SVLDALGTAIHDGGLWGTAEPYPRYTCDYGTPK
TH_2636|M.thermoresistible__bu ALNALFPSHRT------SVLDALGAAMHDGGLWGXXXP-PTICCNARRTR
Mb0180|M.bovis_AF2122/97 GVEQLLVFLPQGAAIAQTVLTPTPGAAQLPLAPAINYPPPCLTGFLPASE
Rv0174|M.tuberculosis_H37Rv GVEQLLVFLPQGAAIAQTVLTPTPGAAQLPLAPAINYPPPCLTGFLPASE
MLBr_02594|M.leprae_Br4923 GVEQMLVFLPQGASIAQTASATFPNMAALDLALSINQPPPCLTGFIPAAE
.:. :: :. . . * .
MMAR_3869|M.marinum_M YLPSAADYPEPFMYT-YCRD--DDPAVSIRGAKNAPRPGGDDTAGPPPG-
MUL_3801|M.ulcerans_Agy99 YLPSAADYPEPFMYT-YCRD--DDPAVSIRGAKNAPRPGGDDTAGPPPG-
MAV_2383|M.avium_104 HASSAADYYEPFMYT-YCRD--DDPAVSIRGAKNAPRPGGDDTAGPPPG-
MAB_1005c|M.abscessus_ATCC_199 LPGSAADYPEPFLYANYCAD--EDPAVLIRGAKNAPRPAGDDTAGPPMG-
Mflv_0005|M.gilvum_PYR-GCK RPPSSADFPEPFMYN-YCRD--DHPGVLVRGAKNAPRPAGDDTAGPPPG-
Mvan_0964|M.vanbaalenii_PYR-1 LPPSSADYPEAFMYT-YCRD--DHPGVLVRGAKNAPRPAGDDTAGPPAG-
MSMEG_1148|M.smegmatis_MC2_155 LPPSSADFPEPFMYT-YCRD--VHPGVLVRGAKNAPRPADDDTAGPPPG-
TH_2636|M.thermoresistible__bu PPPI----------------------------------------------
Mb0180|M.bovis_AF2122/97 WRSPADTSPRPLPSGTYCKIPQDAQLQ-VRGARNIPCVDVPGKRAATPKE
Rv0174|M.tuberculosis_H37Rv WRSPADTSPRPLPSGTYCKIPQDAQLQ-VRGARNIPCVDVLGKRAATPKE
MLBr_02594|M.leprae_Br4923 WRSPADTSLQPLPVGTYCKIPMDTPANSVRGSRNIPCVDVPGKRAASPRE
MMAR_3869|M.marinum_M -------------------------ADLGRRTDPTPRGRFTIPTPYGGPV
MUL_3801|M.ulcerans_Agy99 -------------------------ADLGRRTDPTPRGRFTIPTPYGGPV
MAV_2383|M.avium_104 -------------------------ADLGRRTDPTPRGRYTIPTPYGGPQ
MAB_1005c|M.abscessus_ATCC_199 -------------------------ADMGKKADPTPKGRFQVPTPYGGPT
Mflv_0005|M.gilvum_PYR-GCK -------------------------ADLGRQTDPTPRGRYTIPTPYGGPT
Mvan_0964|M.vanbaalenii_PYR-1 -------------------------ADLGRTTDPTPKGRYTIPTPYGGPT
MSMEG_1148|M.smegmatis_MC2_155 -------------------------ADLGRQTDPTPRGRYTIPTPYGGPT
TH_2636|M.thermoresistible__bu --------------------------------------------------
Mb0180|M.bovis_AF2122/97 CRSKDPYVPLGTNPWFGDPNQILTCPAPGARCDQPVKPGLVIPAPSINTG
Rv0174|M.tuberculosis_H37Rv CRSKDPYVPLGTNPWFGDPNQILTCPAPGARCDQPVKPGLVIPAPSINTG
MLBr_02594|M.leprae_Br4923 CRDSKPYVPLGTNPWFGDPNQLLTCPAPGARCDQPVKPGLVIPAPTVDNG
MMAR_3869|M.marinum_M LPIEPPH-------------------------------------------
MUL_3801|M.ulcerans_Agy99 LPIEPPH-------------------------------------------
MAV_2383|M.avium_104 LPIEPPH-------------------------------------------
MAB_1005c|M.abscessus_ATCC_199 LPIEPPH-------------------------------------------
Mflv_0005|M.gilvum_PYR-GCK LPIEPPR-------------------------------------------
Mvan_0964|M.vanbaalenii_PYR-1 LPIEPPR-------------------------------------------
MSMEG_1148|M.smegmatis_MC2_155 LPIEPPR-------------------------------------------
TH_2636|M.thermoresistible__bu --------------------------------------------------
Mb0180|M.bovis_AF2122/97 LNPAPADQVQGTPPPVSDPLQRPGSGTVQCNGQQPNPCVYTPTSGPSAVY
Rv0174|M.tuberculosis_H37Rv LNPAPADQVQGTPPPVSDPLQRPGSGTVQCNGQQPNPCVYTPTSGPSAVY
MLBr_02594|M.leprae_Br4923 LNPAPADRLPGTPPPVSDPLSQPRSGSVQCNGQQPNTCVYTPSGPPSAVY
MMAR_3869|M.marinum_M ------------------------------------
MUL_3801|M.ulcerans_Agy99 ------------------------------------
MAV_2383|M.avium_104 ------------------------------------
MAB_1005c|M.abscessus_ATCC_199 ------------------------------------
Mflv_0005|M.gilvum_PYR-GCK ------------------------------------
Mvan_0964|M.vanbaalenii_PYR-1 ------------------------------------
MSMEG_1148|M.smegmatis_MC2_155 ------------------------------------
TH_2636|M.thermoresistible__bu ------------------------------------
Mb0180|M.bovis_AF2122/97 SPASGELVGPDGVKYAVANSSTTGDDGWKEMLAPAS
Rv0174|M.tuberculosis_H37Rv SPASGELVGPDGVKYAVANSSTTGDDGWKEMLAPAS
MLBr_02594|M.leprae_Br4923 SPQSGELVGPDGVAYAVENSTKTGDDGWKEMLAPAG