For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VITPIANAVVRVVRAGHRQQTWLSVAGLVLTLVVAAAYLLVGALRLTPFASSYRVTVSLPESGGLLPNQD VALRGVRIGRIESLHITESGVDAIASITSQVKIPASSAVHVSALSPAGEQFLDFEPESDTGPYLGEGAVI GVDQARVPVSLAQLLGDANGLLAQIDPHQVELIKKELSLSQAGPKKLAAIVDGGTFLLSTLDSVLPETTS IIRTGRVVLNLVADKNAGLAATAVALDRTLAGVARMQGGYRSLTERTPHTLAAIDHLFADNSDTMVQLLG SMATAAQLLYLRVPALNALFPDYRGSVLDALASAFHDHGVWATAELYPRYTCDYGTPSYLPSAADYPEPF MYTYCRDDDPAVSIRGAKNAPRPGGDDTAGPPPGADLGRRTDPTPRGRFTIPTPYGGPVLPIEPPH
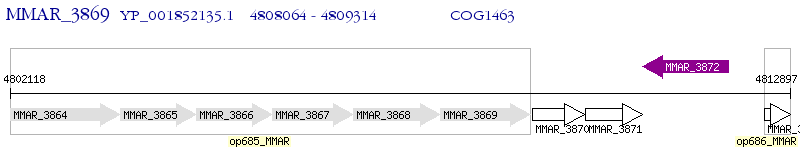
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_3869 | mce5F | - | 100% (416) | Mce family protein Mce5F |
| M. marinum M | MMAR_0182 | mce6F | e-150 | 64.57% (398) | MCE-family protein Mce6F |
| M. marinum M | MMAR_4056 | mce3F_1 | 1e-29 | 26.77% (381) | MCE-family protein Mce3F_1 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0180 | mce1F | 4e-20 | 23.58% (369) | MCE-family protein MCE1F |
| M. gilvum PYR-GCK | Mflv_0005 | - | 1e-166 | 67.95% (415) | virulence factor Mce family protein |
| M. tuberculosis H37Rv | Rv0174 | mce1F | 4e-20 | 23.58% (369) | MCE-family protein MCE1F |
| M. leprae Br4923 | MLBr_02594 | mce1F | 4e-21 | 23.70% (422) | putative secreted protein |
| M. abscessus ATCC 19977 | MAB_4511c | - | 1e-163 | 67.31% (416) | putative Mce family protein |
| M. avium 104 | MAV_2383 | - | 0.0 | 79.76% (415) | mce related protein |
| M. smegmatis MC2 155 | MSMEG_1148 | - | 1e-160 | 66.19% (417) | mce related protein |
| M. thermoresistible (build 8) | TH_2636 | - | 1e-112 | 64.49% (321) | PUTATIVE mce related protein |
| M. ulcerans Agy99 | MUL_3801 | mce5F | 0.0 | 98.08% (416) | Mce family protein Mce5F |
| M. vanbaalenii PYR-1 | Mvan_0964 | - | 1e-163 | 66.27% (415) | hypothetical protein Mvan_0964 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_3869|M.marinum_M MITPIA---NAVVRVVRAGHRQQTWLSVAGLVLTLVVAAAYLLVGALRLT
MUL_3801|M.ulcerans_Agy99 MITPIA---NAVVRVVRAGHRQQTWLSVAGLVLTLVVAAAYLLVGALRLT
MAV_2383|M.avium_104 MIRAVAGFANLVVRAVRTGHRQQVWLSVAGLVLILVVATAYLLIGALRVT
MSMEG_1148|M.smegmatis_MC2_155 MINAVA---DRIVGLVRFGYRKRSWLSALGLVMTLVVATAYLFFGALQVN
TH_2636|M.thermoresistible__bu MIGAVA---DRIVALVRFGHRRRATLSTIGLILTLVVATAYLLFGALQVN
Mflv_0005|M.gilvum_PYR-GCK MINALA---DSVVRVVRTVHRGRAWLSAGALVFILVVAAAYLFVGALQVN
Mvan_0964|M.vanbaalenii_PYR-1 MIGSVA---DAVVGVARFGYRRRAWLSTAALIMTLVVASAYLFFGALQVN
MAB_4511c|M.abscessus_ATCC_199 MIDAVA---NAVVAAVRAGHRQKSMLSTLALVLTLVLAVTYLCVGALRAN
Mb0180|M.bovis_AF2122/97 -------------MLTRFIRRQLILFAIVSVVAIVVLGWYYLRIPSLVG-
Rv0174|M.tuberculosis_H37Rv -------------MLTRFIRRQLILFAIVSVVAIVVLGWYYLRIPSLVG-
MLBr_02594|M.leprae_Br4923 -------------MLTPLIRRQLVLFAILTVISLTVLGVYYLQIPSLVG-
. * :: :: *:. ** . :*
MMAR_3869|M.marinum_M PFASSYRVTVSLPESGGLLPNQDVALRGVRIGRIESLHITESGVDAIASI
MUL_3801|M.ulcerans_Agy99 PFASSYRVTVSLPESGGLLPNQDVALRGVRIGRIESLHLTESGVDAIASI
MAV_2383|M.avium_104 PFASSYRVTVQLPESGGLLPNQDVALRGVRIGRIESLQITDNGVNAVAAI
MSMEG_1148|M.smegmatis_MC2_155 PLDSDYGLTVQLPESAGLLPDQDVTLRGVPIGRVERLDITPSGVNAIVKV
TH_2636|M.thermoresistible__bu PFASSYQVTVALPESGGLLPNQAVTLRGVPVGRVERLDITPEGVNAVVDI
Mflv_0005|M.gilvum_PYR-GCK PLAKSYRVTVQLPESAGLLPNQNVTLRGVPIGRVERLDITPAGVDAVVSM
Mvan_0964|M.vanbaalenii_PYR-1 PLASSYRVTVELPESAGLLPNQNVTMRGVPIGRVERLDITPAGVNAVVSV
MAB_4511c|M.abscessus_ATCC_199 PFASTYRVTVKLSESGGLLPRQDVALRGVKIGTVQSLRITPQGVEATAEI
Mb0180|M.bovis_AF2122/97 --IGQYTLKADLPASGGLYPTANVTYRGITIGKVTAVEPTDQGARVTMSI
Rv0174|M.tuberculosis_H37Rv --IGQYTLKADLPASGGLYPTANVTYRGITIGKVTAVEPTDQGARVTMSI
MLBr_02594|M.leprae_Br4923 --IGRYSLKAELSATGGLYPTANVTYRGIAIGKVTDVEPTATGAEATMSI
* :.. *. :.** * *: **: :* : : * *. . :
MMAR_3869|M.marinum_M TSQVKIPASSAVHVSALSPAGEQFLDFE--PESDTGPYLGEGAVIGVDQA
MUL_3801|M.ulcerans_Agy99 TSQVKIPASSTVHVSALSPAGEQFLDFE--PESDTGPYLSEGAVIGVDQA
MAV_2383|M.avium_104 TSKVRIPANTVAHVSALSPAGEQFINFE--AASDAGPYLHDGSFITSERT
MSMEG_1148|M.smegmatis_MC2_155 KSTVRIPEASDVRVSGLSPAGEQYIDFIDVPAEDSSRFLADGAVIRQGQA
TH_2636|M.thermoresistible__bu DSAVKIPDSSAVRVSGLSPAGEQYIDFI--PEEDTGRYLADGAHIGQGTA
Mflv_0005|M.gilvum_PYR-GCK DSSVAVPMSSAVRVSGLSPAGEQYIDFV--ADSDAGPYLENGSVIAQDNT
Mvan_0964|M.vanbaalenii_PYR-1 DSSAAVPVASSVRVSGLSPAGEQYIDFV--ADSGDGPYLADGAVVSQDNT
MAB_4511c|M.abscessus_ATCC_199 LSKYKIPAAAKVRVAGLSPAGEQYINFEG-TPGQHGPYLSDGSVIAEG-V
Mb0180|M.bovis_AF2122/97 ASNYKIPVDASANVHSVSAVGEQYIDLVS--TGAPGKYFSSGQTITKG--
Rv0174|M.tuberculosis_H37Rv ASNYKIPVDASANVHSVSAVGEQYIDLVS--TGAPGKYFSSGQTITKG--
MLBr_02594|M.leprae_Br4923 DSRYKIPIDSAANVHSVSAVGEQYLDLVS--SGNPGKYLAPGQTITKS--
* :* : ..* .:*..***:::: . :: * :
MMAR_3869|M.marinum_M RVPVSLAQLLGDANGLLAQIDPHQVELIKKELSLSQAGP-KKLAAIVDGG
MUL_3801|M.ulcerans_Agy99 RVPVSLAQLLGDANGLRAQIDPHQVELIKKVLSLSQAGP-KKLAAIVDGG
MAV_2383|M.avium_104 TVPVSLAQLLGDADGLLAQVDPRKIELIKKELSLSKEGP-AKLTAIVDGG
MSMEG_1148|M.smegmatis_MC2_155 TVPVSLADLLADADGALGQVDTAKIELIKRELSLTDAGP-QKLADIVDGG
TH_2636|M.thermoresistible__bu TVPVPLAELLADADGALAQADVEKLEIIRRELSLSPAGP-QKLKDIVDGG
Mflv_0005|M.gilvum_PYR-GCK TVPVSLADLLANADGALSQVDPGKLEVIKHELSMTEAGP-QKLADIIDGG
Mvan_0964|M.vanbaalenii_PYR-1 VVPVSLADLLANADGALAQADPAKLELIKRELSMSEAGP-QKLADIIDGG
MAB_4511c|M.abscessus_ATCC_199 QVPVTLAKLLADADGVLAQVDPKKLELIKKELGLTKEGP-RKLTEIIDGG
Mb0180|M.bovis_AF2122/97 TVPSEIGPALDNSNRGLAALPTEKIGLLLDETAQAVGGLGPALQRLVDST
Rv0174|M.tuberculosis_H37Rv TVPSEIGPALDNSNRGLAALPTEKIGLLLDETAQAVGGLGPALQRLVDST
MLBr_02594|M.leprae_Br4923 KIPIEIGPALDSVNRGLAALPKEKIGSLLDETAQAVGGLGPALQRLVSST
:* :. * . : . :: : . : * * ::..
MMAR_3869|M.marinum_M TFLLSTLDSVLPETTSIIRTGRVVLNLVADKNAGLAATAVALDRTLAGVA
MUL_3801|M.ulcerans_Agy99 TFLLSTLDSVLPETTSIIRTSRVVLNLIADKNAGLAATAVTLDRTLAGVA
MAV_2383|M.avium_104 LFLLSTLDSVLPETTSIIKTSRVVLNLASDKNNGLGAAATELNHTLTGVA
MSMEG_1148|M.smegmatis_MC2_155 TFLLSTLDSVLPETSSLLRTSRVVFTLMADKNPGISVASDNLTETFTGMD
TH_2636|M.thermoresistible__bu TFLLSTLDSVLPETSSLLRTSRVVFTLIDDKNAGIDVAADNLADTFDGVN
Mflv_0005|M.gilvum_PYR-GCK TFLLTTLDSVLPETSSMLRTSRVVFTLAADKNAGIEVASDNLAQTFDGVN
Mvan_0964|M.vanbaalenii_PYR-1 TFLLTTLDSVLPETSSMLRTSRVVLTLAADKNAGIEVASQNLDQVFDGVN
MAB_4511c|M.abscessus_ATCC_199 TFLLSTLDSVLPETTSILKNSRVVLTLAADKNAGIKATADGLGRTLRGVE
Mb0180|M.bovis_AF2122/97 QAIVGDFKTNIGDVNDIIENSGPILDSQVNTGDQIERWARKLNNLAAQTA
Rv0174|M.tuberculosis_H37Rv QAIVGDFKTNIGDVNDIIENSGPILDSQVNTGDQIERWARKLNNLAAQTA
MLBr_02594|M.leprae_Br4923 QAIVDAFKTNITDVNDIIQNSGPILDSQVKSDNAIERWVHNLNVLAAQSA
:: :.: : :...::... :: ... : *
MMAR_3869|M.marinum_M RMQGGYRSLTERTPHTLAAIDHLFADNSDTMVQLLGSMATAAQLLY----
MUL_3801|M.ulcerans_Agy99 RMQGGYRSLTERTPHTLAAIDHLFADNSDTMVQLLGSMATAAQLLY----
MAV_2383|M.avium_104 RMQAGYRRLTSQTPQTLSAVDNLFADNSDTMVQLLGSMATMSQLLY----
MSMEG_1148|M.smegmatis_MC2_155 KMREGYRRLTNQTPETLAAVDNLFVDNSDTMVQLLGNLTTASRLLY----
TH_2636|M.thermoresistible__bu RMREGYRRLTDQTPEVLAHVDNLFADNSDTMVQLLGNLTTTSRLLY----
Mflv_0005|M.gilvum_PYR-GCK RMRDGFRRLTDRAPGTLNSVDNLFADNSDTMVQLLGNLTTTSQLLY----
Mvan_0964|M.vanbaalenii_PYR-1 RMRQGFRTLTEQTPETLAEVDNLFADNSETMVQLLGSLTSTSQLLY----
MAB_4511c|M.abscessus_ATCC_199 KMQNGYRTLIDRTPKTLSAIDNLFSDNSDTMVQLLASLATTSQMLY----
Mb0180|M.bovis_AF2122/97 TRDQNVRSILSQAAPTADEVNAVFSGVRDSLPQTLANLEVVFDMLKRYHA
Rv0174|M.tuberculosis_H37Rv TRDQNVRSILSQAAPTADEVNAVFSGVRDSLPQTLANLEVVFDMLKRYHA
MLBr_02594|M.leprae_Br4923 RQDQHLKSILSQAAPTADEVNTVFSDVRDSLPQTLANFEILIDMLKRYHH
: : .::. . :: :* . ::: * *..: :*
MMAR_3869|M.marinum_M ------------------------------------LRVPALNALFPDYR
MUL_3801|M.ulcerans_Agy99 ------------------------------------LRVPALNALFPDYR
MAV_2383|M.avium_104 ------------------------------------LRVPALNALFPDYR
MSMEG_1148|M.smegmatis_MC2_155 ------------------------------------LRVPALNALFPSYR
TH_2636|M.thermoresistible__bu ------------------------------------LRVPALNALFPSHR
Mflv_0005|M.gilvum_PYR-GCK ------------------------------------LRVPALHALFPDYR
Mvan_0964|M.vanbaalenii_PYR-1 ------------------------------------LRVPALNALFPAHR
MAB_4511c|M.abscessus_ATCC_199 ------------------------------------VRTPAINAFFPNYR
Mb0180|M.bovis_AF2122/97 GVEQLLVFLPQGAAIAQTVLTPTPGAAQLPLAPAINYPPPCLTGFLPASE
Rv0174|M.tuberculosis_H37Rv GVEQLLVFLPQGAAIAQTVLTPTPGAAQLPLAPAINYPPPCLTGFLPASE
MLBr_02594|M.leprae_Br4923 GVEQMLVFLPQGASIAQTASATFPNMAALDLALSINQPPPCLTGFIPAAE
*.: .::* .
MMAR_3869|M.marinum_M GSVLDALASAFHDHGVWATAELYPRYTCDYGTPSYLPSAADYPEPFMYTY
MUL_3801|M.ulcerans_Agy99 GSVLDALASAFHDHGVWATAELYPRYTCDYGTPSYLPSAADYPEPFMYTY
MAV_2383|M.avium_104 GSVLDAVTSVFHDHGVWATADLYPRYVCDYGTPAHASSAADYYEPFMYTY
MSMEG_1148|M.smegmatis_MC2_155 TSVLDALGTAIHDGGLWGTAEPYPRYTCDYGTPKLPPSSADFPEPFMYTY
TH_2636|M.thermoresistible__bu TSVLDALGAAMHDGGLWGXXXP-PTICCNARRTRPPPI------------
Mflv_0005|M.gilvum_PYR-GCK TSVLDAIGSAMHDNGLWATGEIYPRYTCDYGTPRRPPSSADFPEPFMYNY
Mvan_0964|M.vanbaalenii_PYR-1 TSVLDAIGSIMHDNGLWATGDIYPRYSCDYGTPRLPPSSADYPEAFMYTY
MAB_4511c|M.abscessus_ATCC_199 GSTFGAIADTMRDNGIWVTADLYPRYACDYGTPRVPPSSADFPEPPVYTY
Mb0180|M.bovis_AF2122/97 WRSPADTSPRPLPSGTYCKIPQDAQLQ-VRGARNIPCVDVPGKRAATPKE
Rv0174|M.tuberculosis_H37Rv WRSPADTSPRPLPSGTYCKIPQDAQLQ-VRGARNIPCVDVLGKRAATPKE
MLBr_02594|M.leprae_Br4923 WRSPADTSLQPLPVGTYCKIPMDTPANSVRGSRNIPCVDVPGKRAASPRE
* : .
MMAR_3869|M.marinum_M CRDDDPAVSIRG----------------AKNAPRPGGDDTAGPPPGADLG
MUL_3801|M.ulcerans_Agy99 CRDDDPAVSIRG----------------AKNAPRPGGDDTAGPPPGADLG
MAV_2383|M.avium_104 CRDDDPAVSIRG----------------AKNAPRPGGDDTAGPPPGADLG
MSMEG_1148|M.smegmatis_MC2_155 CRDVHPGVLVRG----------------AKNAPRPADDDTAGPPPGADLG
TH_2636|M.thermoresistible__bu --------------------------------------------------
Mflv_0005|M.gilvum_PYR-GCK CRDDHPGVLVRG----------------AKNAPRPAGDDTAGPPPGADLG
Mvan_0964|M.vanbaalenii_PYR-1 CRDDHPGVLVRG----------------AKNAPRPAGDDTAGPPAGADLG
MAB_4511c|M.abscessus_ATCC_199 CADDDPAVLIRG----------------AKNAPRPAGDDTAGPPPGADLG
Mb0180|M.bovis_AF2122/97 CRSKDPYVPLGTNPWFGDPNQILTCPAPGARCDQPVKPGLVIPAPSINTG
Rv0174|M.tuberculosis_H37Rv CRSKDPYVPLGTNPWFGDPNQILTCPAPGARCDQPVKPGLVIPAPSINTG
MLBr_02594|M.leprae_Br4923 CRDSKPYVPLGTNPWFGDPNQLLTCPAPGARCDQPVKPGLVIPAPTVDNG
MMAR_3869|M.marinum_M RRTDPTPRGRFTIP--------------------------TPYGGPVLPI
MUL_3801|M.ulcerans_Agy99 RRTDPTPRGRFTIP--------------------------TPYGGPVLPI
MAV_2383|M.avium_104 RRTDPTPRGRYTIP--------------------------TPYGGPQLPI
MSMEG_1148|M.smegmatis_MC2_155 RQTDPTPRGRYTIP--------------------------TPYGGPTLPI
TH_2636|M.thermoresistible__bu --------------------------------------------------
Mflv_0005|M.gilvum_PYR-GCK RQTDPTPRGRYTIP--------------------------TPYGGPTLPI
Mvan_0964|M.vanbaalenii_PYR-1 RTTDPTPKGRYTIP--------------------------TPYGGPTLPI
MAB_4511c|M.abscessus_ATCC_199 KKTDPTPKGRFTIP--------------------------TPPGGPTLPI
Mb0180|M.bovis_AF2122/97 LNPAPADQVQGTPPPVSDPLQRPGSGTVQCNGQQPNPCVYTPTSGPSAVY
Rv0174|M.tuberculosis_H37Rv LNPAPADQVQGTPPPVSDPLQRPGSGTVQCNGQQPNPCVYTPTSGPSAVY
MLBr_02594|M.leprae_Br4923 LNPAPADRLPGTPPPVSDPLSQPRSGSVQCNGQQPNTCVYTPSGPPSAVY
MMAR_3869|M.marinum_M EPPH--------------------------------
MUL_3801|M.ulcerans_Agy99 EPPH--------------------------------
MAV_2383|M.avium_104 EPPH--------------------------------
MSMEG_1148|M.smegmatis_MC2_155 EPPR--------------------------------
TH_2636|M.thermoresistible__bu ------------------------------------
Mflv_0005|M.gilvum_PYR-GCK EPPR--------------------------------
Mvan_0964|M.vanbaalenii_PYR-1 EPPR--------------------------------
MAB_4511c|M.abscessus_ATCC_199 EPPR--------------------------------
Mb0180|M.bovis_AF2122/97 SPASGELVGPDGVKYAVANSSTTGDDGWKEMLAPAS
Rv0174|M.tuberculosis_H37Rv SPASGELVGPDGVKYAVANSSTTGDDGWKEMLAPAS
MLBr_02594|M.leprae_Br4923 SPQSGELVGPDGVAYAVENSTKTGDDGWKEMLAPAG