For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RFSIWGQTNQSWADLLDIAQYADDGTWRAFYAADHFMSHADTAEERTDFHEATALFAALAAATSRIRLAP LVLSMTFRHPAVLANWAATVDHVSQGRFTLGLGAGWQENEHQRYGLELGKPGARVDRFSEGLQVITGLLN EPVTTVDGQYYRLAEGICEPKPLQRPLPILIGATQPRMLALTARYAHEWNQWSSPGTFREVSGRLDAAAE KIGRDPATIWRSTQARVIVTDGPQSEARAAETAAASPIPVLYGTAERIAEQMAPWTQEGVDEVILPDHLL GSGSARRDAYDALAQALRPLSNETD*
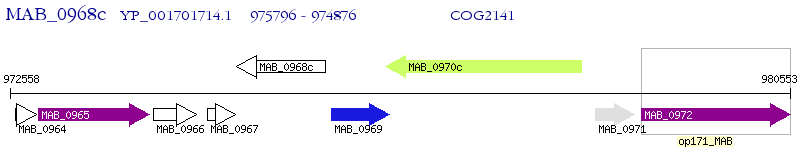
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0968c | - | - | 100% (306) | luciferase-like hypothetical protein |
| M. abscessus ATCC 19977 | MAB_4797 | - | 5e-43 | 42.35% (255) | luciferase-like monooxygenase |
| M. abscessus ATCC 19977 | MAB_4496c | - | 3e-37 | 39.04% (251) | luciferase-like monooxygenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0815c | - | 1e-36 | 33.79% (290) | hypothetical protein Mb0815c |
| M. gilvum PYR-GCK | Mflv_1117 | - | 2e-37 | 41.26% (223) | luciferase family protein |
| M. tuberculosis H37Rv | Rv0791c | - | 1e-36 | 33.79% (290) | hypothetical protein Rv0791c |
| M. leprae Br4923 | MLBr_00348 | - | 8e-13 | 30.09% (216) | putative coenzyme F420-dependent oxidoreductase |
| M. marinum M | MMAR_5411 | - | 4e-39 | 42.22% (225) | hypothetical protein MMAR_5411 |
| M. avium 104 | MAV_0169 | - | 2e-37 | 40.77% (233) | hypothetical protein MAV_0169 |
| M. smegmatis MC2 155 | MSMEG_3301 | - | 2e-37 | 41.33% (225) | hypothetical protein MSMEG_3301 |
| M. thermoresistible (build 8) | TH_4285 | - | 3e-36 | 38.02% (242) | PUTATIVE luciferase family protein |
| M. ulcerans Agy99 | MUL_5034 | - | 3e-39 | 43.11% (225) | hypothetical protein MUL_5034 |
| M. vanbaalenii PYR-1 | Mvan_5695 | - | 2e-37 | 40.44% (225) | luciferase family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1117|M.gilvum_PYR-GCK -----------MRFAFKTSPQNTTWSDMLTVWKAADDIDVFESGWTFDHF
Mvan_5695|M.vanbaalenii_PYR-1 -----------MRFAFKTSPQNTAWSDMLSVWKAADDIDVFESGWTFDHF
TH_4285|M.thermoresistible__bu -----------MQFGFKTSPQNTTWQDMLSVWRAADDIDVFESGWTFDHF
MMAR_5411|M.marinum_M -----------MRFAFKTSPQNTTWEQMLAVWQTADGIDVYESGWTFDHF
MUL_5034|M.ulcerans_Agy99 -----------MRFAFKNSPQNTTWEQMLAVWQTADGIDVYESGWTFDHF
MAV_0169|M.avium_104 -----------MRFAFKTSPQNTTWAQMLAVWQAADDIDVYESGWTFDHF
MSMEG_3301|M.smegmatis_MC2_155 -----------MRFAFKTSPQNTTWQDMLAVWQTADDIEVFESGWTFDHF
Mb0815c|M.bovis_AF2122/97 MNAKDDPHFGLMLAATVNGLAVGSYREMVVVSQTAEEYG-FDSVWLCDHF
Rv0791c|M.tuberculosis_H37Rv MNAKDDPHFGLMLAATVNGLAVGSYREMVVVSQTAEEYG-FDSVWLCDHF
MAB_0968c|M.abscessus_ATCC_199 -----------MRFSIWG-QTNQSWADLLDIAQYADDGT-WRAFYAADHF
MLBr_00348|M.leprae_Br4923 -------------MVERANAMKLGLQLGYWAAQPPGNAAELVAAAEGAGF
: . : *
Mflv_1117|M.gilvum_PYR-GCK YPIFS-----------------------------DPTGPCLEGWVTLTAL
Mvan_5695|M.vanbaalenii_PYR-1 YPIFS-----------------------------DPSGPCLEGWVTLTAL
TH_4285|M.thermoresistible__bu YPIFS-----------------------------DPTGPCLEGWTTLTAL
MMAR_5411|M.marinum_M YPIFS-----------------------------DPTGPCLEGWTTLTAL
MUL_5034|M.ulcerans_Agy99 YPIFS-----------------------------DPTGPCLEGWTTLTAL
MAV_0169|M.avium_104 YPIFS-----------------------------DSSGPCLEGWTTLTAL
MSMEG_3301|M.smegmatis_MC2_155 YPIVG-----------------------------DSSGPCLEGWTTLTAL
Mb0815c|M.bovis_AF2122/97 LTISPGEYAKVAGIAADTGSATGTETGGAGQCAPSRSLPLLECWTALAAL
Rv0791c|M.tuberculosis_H37Rv LTISPGEYAKVAGIAADTGSATGTETGGAGQCAPSRSLPLLECWTALAAL
MAB_0968c|M.abscessus_ATCC_199 MSHADT---------------------------AEERTDFHEATALFAAL
MLBr_00348|M.leprae_Br4923 DAVFTG------------------------------EAWGSDAYTPLAWW
. : . ::
Mflv_1117|M.gilvum_PYR-GCK AQATTRLRVGVLVTGIHYRHPAVLANMASALDIVSGGRLELGIGAGWNEE
Mvan_5695|M.vanbaalenii_PYR-1 AQATTRLRVGVLVTGIHYRHPAVLANMASALDIVSDGRLELGIGAGWNEE
TH_4285|M.thermoresistible__bu AQATTRLRVGVLVTGIHYRHPAVLANMASALDIISAGRLELGIGAGWNEE
MMAR_5411|M.marinum_M AQATTRLRLGTLVTGVHYRHPAVLANMAAALDIISHGRLELGIGAGWNEE
MUL_5034|M.ulcerans_Agy99 AQATTRLRLGTLVTGIHYRHPAVLANMAAALDIISHGRLELGIGAGWNEE
MAV_0169|M.avium_104 AQATKRLRLGTLVTGIHYRHPAVLANMAAAVDIISGGRLELGIGAGWNEE
MSMEG_3301|M.smegmatis_MC2_155 AQATSRLRLGTLVTGVHYRHPAVLANMAAALDIISNGRLELGIGAGWNEE
Mb0815c|M.bovis_AF2122/97 SRDTTKLRLGTSVLCNSYRHPSVLAKMAATLDVISQGRLDLGLGAGWFRR
Rv0791c|M.tuberculosis_H37Rv SRDTTKLRLGTSVLCNSYRHPSVLAKMAATLDVISQGRLDLGLGAGWFRR
MAB_0968c|M.abscessus_ATCC_199 AAATSRIRLAPLVLSMTFRHPAVLANWAATVDHVSQGRFTLGLGAGWQEN
MLBr_00348|M.leprae_Br4923 GSSTQRVRLGTSVVQLSARTPTACAMAALTLDHLSGGRHILGLGVSGPQV
. * ::*:. * * *:. * * ::* :* ** **:*.. .
Mflv_1117|M.gilvum_PYR-GCK ESGAYGIELGSITERFDRFEEACQVLRGLLSQETTTS-DGRFYQLKDARN
Mvan_5695|M.vanbaalenii_PYR-1 ESGAYGIELGSIKERFDRFEEACQVLKGLLSQDTTTF-DGKFYQLKDARN
TH_4285|M.thermoresistible__bu ESGAYGIELGTLKERFDRFEEACQVLISLLSQETTDF-DGAYYRLKSARN
MMAR_5411|M.marinum_M ESSAYGIELGSVKERFDRFEEACAVLTGLLSQPSTTF-DGKYYQLEDARN
MUL_5034|M.ulcerans_Agy99 ESGAYGIELGSVKERFDRFEEACAVLTGLLSQPSTTF-DGKYYQLEDARN
MAV_0169|M.avium_104 ESGAYGIELGSIRERFDRFEEACAVLTSLLSQETTDF-DGKFYQLKGARN
MSMEG_3301|M.smegmatis_MC2_155 ESGAYGIELGSIKERLDRFEEATQVLIGLLSQDSTDF-DGAFYTLKNARN
Mb0815c|M.bovis_AF2122/97 ESQAYGIPFPPVGDRVSALAESLQVIKAVWTEPNPTY-AGRFYTLDGATC
Rv0791c|M.tuberculosis_H37Rv ESQAYGIPFPPVGDRVSALAESLQVIKAVWTEPNPTY-AGRFYTLDGATC
MAB_0968c|M.abscessus_ATCC_199 EHQRYGLELGKPGARVDRFSEGLQVITGLLNEPVTTV-DGQYYRLAEGIC
MLBr_00348|M.leprae_Br4923 VEGWYGQPFPKP---LSRTREYIDIVRQVWAREAPVVSDGQHYPLPLTGA
** : .. * :: : . . * .* *
Mflv_1117|M.gilvum_PYR-GCK EPKGPQQPHPPI---------CIGGNGEKRTLRITAQYADHWNYVGGTPE
Mvan_5695|M.vanbaalenii_PYR-1 EPKGPQRPHPPI---------CIGGSGEKRTLRITAQYADHWNFVGGPPE
TH_4285|M.thermoresistible__bu EPKGPQQPHPPI---------CIGGSGEKRTLRITAKYADHWNFVGGPPE
MMAR_5411|M.marinum_M EPKGPQRPHPPI---------CIGGSGEKRTLRITARYAQHWNFAGGTPE
MUL_5034|M.ulcerans_Agy99 EPKGPQRPHSPI---------CIGGSGEKRTLRITARYAQHWNFAGGTPE
MAV_0169|M.avium_104 EPKGPQRPHPPI---------CIGGSGEKRTLRITARYAQHWNFVGGPPD
MSMEG_3301|M.smegmatis_MC2_155 EPKGPQRPHPPI---------VMGGSGEKRTLRITAKYADHWNFVLGTPE
Mb0815c|M.bovis_AF2122/97 DPPPVQRPHPPL---------WIGGEGD-RVQRIAAKHAQGLNVRWWSPQ
Rv0791c|M.tuberculosis_H37Rv DPPPVQRPHPPL---------WIGGEGD-RVQRIAAKHAQGLNVRWWSPQ
MAB_0968c|M.abscessus_ATCC_199 EPKPLQRPLP-----------ILIGATQPRMLALTARYAHEWN-QWSSPG
MLBr_00348|M.leprae_Br4923 GATGLGKALKPITHPLRADIPIMLGAEGPKNVALAAEICDGWLPIFFSPR
. :. : * : ::*. .. .*
Mflv_1117|M.gilvum_PYR-GCK EFARKRDVLAAHCADIG--RDPKEIMLSAHVRLGADLD------------
Mvan_5695|M.vanbaalenii_PYR-1 EFARKRDVLARHCADIG--RDPGEISLSAHIRLGADLD------------
TH_4285|M.thermoresistible__bu EFARKRDVLAAHCADVG--RDPREIRLSAHVRLGEDGD------------
MMAR_5411|M.marinum_M EFARKRDVLAAHCADIG--RDPKEITLSAHLGLAPERD------------
MUL_5034|M.ulcerans_Agy99 EFARKRDVLAAHCADIG--RDPKEITLSAHRGLAPERD------------
MAV_0169|M.avium_104 VFAHKRDVLASHCADIG--RDPKEILLSAHLRLEPDLD------------
MSMEG_3301|M.smegmatis_MC2_155 EFAHKRDVLWSHCADIG--RDPKEITLSAHVWHNAEHD------------
Mb0815c|M.bovis_AF2122/97 QVTQRRGFLTQASEAAG--RDPDTLRLSVTLLLAPTQSGEEEVRIREEFA
Rv0791c|M.tuberculosis_H37Rv QVTQRRGFLTQASEAAG--RDPDTLRLSVTLLLAPTQSGEEEVRIREEFA
MAB_0968c|M.abscessus_ATCC_199 TFREVSGRLDAAAEKIG--RDPATIWRSTQARVIVTDGPQSEARAAETAA
MLBr_00348|M.leprae_Br4923 MADMYNEWLDEGFARTGARRSREDFEICASAQIVVTEDRAAAFAGIKPFL
* * *. : .. .
Mflv_1117|M.gilvum_PYR-GCK -----------VDKVIAEAAALGDEGLDLAIVYLPPPYDPAVLEPLAEKI
Mvan_5695|M.vanbaalenii_PYR-1 -----------YDKVIEEAAALGAEGLELAIVYLPLPHDPAVLEPLADKI
TH_4285|M.thermoresistible__bu -----------CRKVVDEAAALASEGLDLAIVYLPTPHTPAVLEPLAEAI
MMAR_5411|M.marinum_M -----------FGQVIENAAALAAEGLDLGIVYIAPPHDPAVLEPMAEAI
MUL_5034|M.ulcerans_Agy99 -----------FGQVIENAAALAAEGLDLGIVYIAPPHDPAVLEPMAEAI
MAV_0169|M.avium_104 -----------YGKVIDDAAALAAEGLDLGIVYLPPPHDPAVLEPLAEAI
MSMEG_3301|M.smegmatis_MC2_155 -----------HQKLLTEVAAFGEVGLDLAIIYLPPPLTPAILEPLAAAI
Mb0815c|M.bovis_AF2122/97 SIPEPGLIVGTPDRCVERIREYQDRGVGHFLFTIPHVVKSDYLHIIGSDI
Rv0791c|M.tuberculosis_H37Rv SIPEPGLIVGTPDRCVERIREYQDRGVGHFLFTIPHVVKSDYLHIIGSDI
MAB_0968c|M.abscessus_ATCC_199 ASPIPVLYG-TAERIAEQMAPWTQEGVDEVILPDHLLGSGSARRDAYDAL
MLBr_00348|M.leprae_Br4923 ALYMGGMG---AEGTNFHADVYRRMGYAEVVDEVTALFRSNQKDKAAEII
* : :
Mflv_1117|M.gilvum_PYR-GCK RASGLLDSNVDG-------------------------------------
Mvan_5695|M.vanbaalenii_PYR-1 RASGLLDSKAR--------------------------------------
TH_4285|M.thermoresistible__bu RDSGLPAR-----------------------------------------
MMAR_5411|M.marinum_M RESGLIDG-----------------------------------------
MUL_5034|M.ulcerans_Agy99 RESGLIDG-----------------------------------------
MAV_0169|M.avium_104 RDSGLLRS-----------------------------------------
MSMEG_3301|M.smegmatis_MC2_155 TASGLLTTTKE--------------------------------------
Mb0815c|M.bovis_AF2122/97 IPRVKTEVTIP--------------------------------------
Rv0791c|M.tuberculosis_H37Rv IPRVKTEVTIP--------------------------------------
MAB_0968c|M.abscessus_ATCC_199 AQALRPLSNETD-------------------------------------
MLBr_00348|M.leprae_Br4923 PNELVDSTMIVGDVDYVCKQIAAWSAAGVTMMMVSASSVEQVRDLAGLF