For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TTPRTRPSRQVGRPGYDLDSLLAVAVSVFNQRGYDGTSMEHLSTRLGISKSSIYHHVSGKEELLRLAVDR ALDALFAATEAPDTTTGPAIDRLENLVKRSIQVLVQELPYVTLLLRIRGNTAAERRALARRREFDDIVGN LVEEACNEGSIRAGVDPALTSRLIFGTVNSLIEWYRPSRTLSADEVADAVATMIFDGLRTRSGQH*
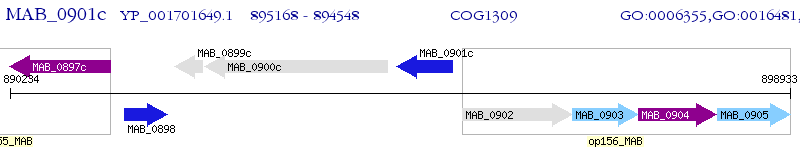
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0901c | - | - | 100% (206) | TetR family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_4912c | - | 4e-11 | 25.50% (200) | TetR family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_1149 | - | 4e-11 | 24.24% (198) | TetR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3587c | - | 2e-13 | 27.59% (203) | TetR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_4584 | - | 3e-12 | 27.68% (177) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv3557c | - | 2e-13 | 27.59% (203) | TetR family transcriptional regulator |
| M. leprae Br4923 | MLBr_00717 | - | 1e-07 | 33.73% (83) | putative TetR-family transcriptional regulator |
| M. marinum M | MMAR_5046 | - | 2e-12 | 26.63% (199) | TetR family transcriptional regulator |
| M. avium 104 | MAV_0603 | - | 3e-13 | 26.98% (189) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_6009 | - | 4e-14 | 26.63% (199) | transcriptional regulator, TetR family protein |
| M. thermoresistible (build 8) | TH_3291 | - | 2e-14 | 27.81% (187) | PUTATIVE transcriptional regulator, TetR family protein |
| M. ulcerans Agy99 | MUL_4635 | - | 4e-12 | 24.86% (181) | TetR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_0318 | - | 4e-14 | 27.96% (186) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3587c|M.bovis_AF2122/97 -------MDRVAGQVNSRRGELLELAAAMFAERGLRATTVRDIADGAGIL
Rv3557c|M.tuberculosis_H37Rv -------MDRVAGQVNSRRGELLELAAAMFAERGLRATTVRDIADGAGIL
MMAR_5046|M.marinum_M -------MERMTGQANSRRGELLGLAAVMFAERGLRATTVRDIADGAGIL
MAV_0603|M.avium_104 ----------MTGPANSRRDELLELAAAMFAERGLRATTVRDIADSAGIL
MSMEG_6009|M.smegmatis_MC2_155 ------MAPDTPSQPASRRDELLQLAATMFADRGLKATTVRDIADSAGIL
Mflv_4584|M.gilvum_PYR-GCK -------MHDTPSQICDR----HPASADLSAEKGPNDSKFEDVAAVTGVP
Mvan_0318|M.vanbaalenii_PYR-1 -------MRKIPRQISDR----LPAAADLFAEKGLNDSKIEDVAAVTGVP
MAB_0901c|M.abscessus_ATCC_199 MTTPRTRPSRQVGRPGYDLDSLLAVAVSVFNQRGYDGTSMEHLSTRLGIS
TH_3291|M.thermoresistible__bu ------VNDEQESKSAATRARILDAAAHVLSVRGYAGLRLTDVAARADIQ
MUL_4635|M.ulcerans_Agy99 ------------MTGSERRHQLIGIARSLFAERGYDGTSIEEIAQRANVS
MLBr_00717|M.leprae_Br4923 -----MTTARRRLSPEDRRAELLALGAEVFGKRPYDEVRIDEIAERAGVS
. : : . .:: .:
Mb3587c|M.bovis_AF2122/97 SGSLYHHFASKEEMVDELLRGFLDWLFARYRDIVDSTANPLERLQGLFMA
Rv3557c|M.tuberculosis_H37Rv SGSLYHHFASKEEMVDELLRGFLDWLFARYRDIVDSTANPLERLQGLFMA
MMAR_5046|M.marinum_M SGSLYHHFASKEEMVDELLREFLDWLFARYLEIVDGEPNPLERLKGLFLA
MAV_0603|M.avium_104 SGSLYHHFSSKEEMVDEVLRSFLDWLFARYREIIDSESNPLERLKGLFMA
MSMEG_6009|M.smegmatis_MC2_155 SGSLYHHFKSKEQMVEEVLRDFLDWLFGRYQQILDTATSPLEKLTGLFMA
Mflv_4584|M.gilvum_PYR-GCK KATLYYYFAGKEDILAFLLEDLLKDISEAVTAIVHTDGTGAERLELVIRA
Mvan_0318|M.vanbaalenii_PYR-1 KATLYYYFAGKEDILAFLLEDLLKDISEAVTAIVHTDGTGAERLELVIRA
MAB_0901c|M.abscessus_ATCC_199 KSSIYHHVSGKEELLRLAVDRALDALFAATEAPDTTTGPAIDRLENLVKR
TH_3291|M.thermoresistible__bu APAIYYYYPSREALIEEVMWVGIADTRDQVVAALDALPDGTPALDRLLTA
MUL_4635|M.ulcerans_Agy99 KPVVYEHFGGKEGLYAVVVDREMSALLDGITSSLTNNRS-RVRVERVALA
MLBr_00717|M.leprae_Br4923 RALMYHYFPDKRAFFAAVVKDEADRLYAATNIPPPAGLTMFEEVRVGVLA
:* : .:. : : :
Mb3587c|M.bovis_AF2122/97 SFEAIEHHHAQVVIYQDEAQRLASQPRFSYIEDRNKQ--QRKMWVDVLNQ
Rv3557c|M.tuberculosis_H37Rv SFEAIEHHHAQVVIYQDEAQRLASQPRFSYIEDRNKQ--QRKMWVDVLNQ
MMAR_5046|M.marinum_M SFEAIEHRHAQVVIYQDEAQRLSSQPRFSYLEDRNKE--QRKMWVDVLNQ
MAV_0603|M.avium_104 SFEAIEHRHAQVVIYQDEAKRLLSQPRFSYIEDMNRQ--QRKMWLEVLNQ
MSMEG_6009|M.smegmatis_MC2_155 SFEAIEHRHAQVVIYQDEAKRLSDLPQFDFVETRNKE--QRKMWVDILQE
Mflv_4584|M.gilvum_PYR-GCK QLRAMAQRPAVCRALIGELGRAARMP--AIAEMINTA--YQQPVEILLIE
Mvan_0318|M.vanbaalenii_PYR-1 QLRAMAQRPAVCRALIGELGRAARMP--AIAEMINTA--YQQPVETLLVE
MAB_0901c|M.abscessus_ATCC_199 SIQVLVQELPYVTLLLRIRGNTAAERR---ALARRRE--FDDIVGNLVEE
TH_3291|M.thermoresistible__bu VATHLQHALEISDYATAAIRNAGQVPLAIRKRQIREEERYGEVWRRLIND
MUL_4635|M.ulcerans_Agy99 LLTYVEERTDGFRIMIRDSP--ASISSGTYSSLLNDA--VSQVSSILAGD
MLBr_00717|M.leprae_Br4923 YMAYHKQNPEAAWAAYVGLGRSDPVLLGIDDEAKNRQMEHIMSRIGDVVS
. . .
Mb3587c|M.bovis_AF2122/97 GIEEGYFRPDLDVD--LVYRFIRDTTWVSVRWYRPGGPLTAQQVGQQYLA
Rv3557c|M.tuberculosis_H37Rv GIEEGYFRPDLDVD--LVYRFIRDTTWVSVRWYRPGGPLTAQQVGQQYLA
MMAR_5046|M.marinum_M GIEEGYFRPDLDVD--LVYRFIRDTTWVSVRWYKPGGALTAQQVGQKYLA
MAV_0603|M.avium_104 GVDEGYFRPDLDVD--LVYRFIRDTTWVSVRWYQPGGPLTAEQVGQQYLA
MSMEG_6009|M.smegmatis_MC2_155 GVADGSFRPDLDVD--LVYRFIRDTTWVSVRWYKPGGPLSAEQVGQQYLA
Mflv_4584|M.gilvum_PYR-GCK GARDGSLVAQPDARS-TSIALFGAVTINALMYLVTENHLDETVVASTLSA
Mvan_0318|M.vanbaalenii_PYR-1 GARDGSLVAQPDARS-TSIALFGAVTINALMYLVTENHLDETVVARTLSA
MAB_0901c|M.abscessus_ATCC_199 ACNEGSIRAGVDPA--LTSRLIFGTVNSLIEWYRPSRTLSADEVADAVAT
TH_3291|M.thermoresistible__bu LAREGQLRPELDLY--IAQMLILGALNWAAEWWDP-RRSSVETVIANAQT
MUL_4635|M.ulcerans_Agy99 FARRG---LDSELAP-LYAQALVGSVSMTAQWWLDAREPKKEVVAAHLVN
MLBr_00717|M.leprae_Br4923 VVPESKLHPDIERDLRVIVHGWLAFTFEVCRQRIMDPSTDADRLADSCAH
. : :
Mb3587c|M.bovis_AF2122/97 IVLGGITK-EGV----------------
Rv3557c|M.tuberculosis_H37Rv IVLGGITK-EGV----------------
MMAR_5046|M.marinum_M IVLGGIQK-EGV----------------
MAV_0603|M.avium_104 IVLGGITV-DAKKD--------------
MSMEG_6009|M.smegmatis_MC2_155 IVLGGITQSQGDKHA-------------
Mflv_4584|M.gilvum_PYR-GCK VMFDGLRPRTGDQS--------------
Mvan_0318|M.vanbaalenii_PYR-1 VMFDGLRPRTGDQS--------------
MAB_0901c|M.abscessus_ATCC_199 MIFDGLRTRSGQH---------------
TH_3291|M.thermoresistible__bu IIKHGITTVE------------------
MUL_4635|M.ulcerans_Agy99 LVWNGLTHLEADPQLQDE----------
MLBr_00717|M.leprae_Br4923 ALLDAIARVPEISKELADAMAIARQPPQ
: .: