For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSTAVTPSPEMCSTPLELFDTDRRLDQDERDIAATVRKFVDTKLRPNIAEWFESATLPRDLARQFGELGV LGMHLDGYGCAGTNAVSYGLACLELEAGDSGLRSFVSVQGSLSMYSIHRYGSEEQKQEWLPQLASGHAIG CFGLTEPDFGSNPGGMRTRARRDGKDWILDGTKMWITNGNLADVATVWAQTDDGVRGFVVPTATKGFAAN VIHKKLSLRASVTSELVLDGVRLPDSAELPGARGLSAPLSCLNEARFGIVFGALGAARDSLQAALDYTRT REVFGRPLSSYQLSQEKLANMTVQLGMGMLLALHLGRIKDSVGLRPEQVSLGKLNNVREALNIARECRTL LGASGITLEYSPLRHANNLESVLTYEGTSEMHLLSIGRALTGQAAFR
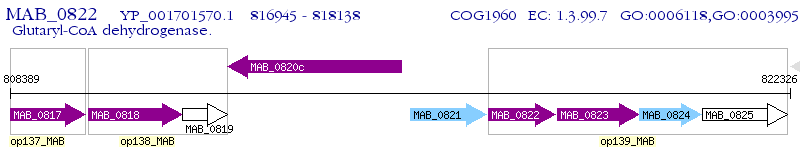
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0822 | - | - | 100% (397) | acyl-CoA dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4609c | - | e-146 | 65.28% (386) | putative acyl-CoA dehydrogenase FadE |
| M. abscessus ATCC 19977 | MAB_4538c | - | 7e-46 | 32.72% (379) | putative acyl-CoA dehydrogenase FadE |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0406c | fadE7 | 1e-142 | 63.16% (399) | acyl-CoA dehydrogenase FADE7 |
| M. gilvum PYR-GCK | Mflv_4513 | - | 0.0 | 80.80% (401) | acyl-CoA dehydrogenase domain-containing protein |
| M. tuberculosis H37Rv | Rv0400c | fadE7 | 1e-142 | 63.16% (399) | acyl-CoA dehydrogenase FADE7 |
| M. leprae Br4923 | MLBr_00737 | fadE25 | 6e-41 | 30.65% (385) | putative acyl-CoA dehydrogenase |
| M. marinum M | MMAR_0698 | fadE7 | 1e-145 | 63.66% (399) | acyl-CoA dehydrogenase FadE7 |
| M. avium 104 | MAV_4767 | - | 1e-145 | 66.05% (377) | putative acyl-CoA dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_6686 | - | 0.0 | 82.78% (395) | glutaryl-CoA dehydrogenase |
| M. thermoresistible (build 8) | TH_2172 | - | 1e-151 | 67.36% (383) | glutaryl-CoA dehydrogenase |
| M. ulcerans Agy99 | MUL_2825 | fadE7 | 1e-145 | 63.66% (399) | acyl-CoA dehydrogenase FadE7 |
| M. vanbaalenii PYR-1 | Mvan_1857 | - | 0.0 | 81.55% (401) | acyl-CoA dehydrogenase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0406c|M.bovis_AF2122/97 ----MSTPTPPALDRDDPLGLDASLSSDEIAVRDTVRRFCAEHVTPHVAA
Rv0400c|M.tuberculosis_H37Rv ----MSTPTPPALDRDDPLGLDASLSSDEIAVRDTVRRFCAEHVTPHVAA
MMAR_0698|M.marinum_M ----MSTETPPTFDPHDPLGLDASLSDDEIAVRDTVRKFCAEHITAHVAE
MUL_2825|M.ulcerans_Agy99 ----MSTETPPTFDPHDPLGLDASLSDDEIAVRDTVRKFCAEHITAHVAE
MAV_4767|M.avium_104 MSTETSTKTPPAFDRYDPLGLDALLSDDERAVRDTVRGFCAEHVLPHVAE
Mflv_4513|M.gilvum_PYR-GCK MSTTTAPATKAVYAPLELFDTARLLEQDEREIAATVRKFVDSELKPNVED
Mvan_1857|M.vanbaalenii_PYR-1 -MTTTAAPQKAVYSPLELFDTARLLDHDEREIAATVRKFVDTELKPNIEG
MSMEG_6686|M.smegmatis_MC2_155 -MTLTAPSKKSTYAPLELFDTDRLLDQDERDIAATVRQFVDTRLKPNVEG
MAB_0822|M.abscessus_ATCC_1997 MSTAVTPSPEMCSTPLELFDTDRRLDQDERDIAATVRKFVDTKLRPNIAE
TH_2172|M.thermoresistible__bu --------------VDDLLDLDALLTPEDIELRDTVRRFGEQRLRPHVAD
MLBr_00737|M.leprae_Br4923 ---------MVGWSGNPLFDLFKLPEEHN-ELRATIRALAEKEIAPHAAD
:. .: : *:* : .: .:
Mb0406c|M.bovis_AF2122/97 WFEDGDLPVARDLAKQFGELGLLGMQL-HGHGCGGASAVHYGLACRELEA
Rv0400c|M.tuberculosis_H37Rv WFEDGDLPVARDLAKQFGELGLLGMQL-HGHGCGGASAVHYGLACRELEA
MMAR_0698|M.marinum_M WFENGDLPAARDLAKQLGGLGLLGMHL-QGYGCGGASAVHYGLACLELEA
MUL_2825|M.ulcerans_Agy99 WFENGDLPAARDLAKQLGGLGLLGMHL-QGYGCGGASAVHYGLACLELEA
MAV_4767|M.avium_104 WFEIGDLP-VRELAREFGRLGLLGMHL-HGYGCGGASAVHYGLACVELEA
Mflv_4513|M.gilvum_PYR-GCK WFESATLP--KELGKRFGELGLLGMHL-QGYGCAGTNAVSYGLACMELEA
Mvan_1857|M.vanbaalenii_PYR-1 WFESSTLP--RELGRRFGELGVLGMHL-EGYGCAGTNAVSYGLACLELEA
MSMEG_6686|M.smegmatis_MC2_155 WFESATLP--SELAKEFGNLGVLGMHL-QGYGCAGTNAVSYGLACMELEA
MAB_0822|M.abscessus_ATCC_1997 WFESATLP--RDLARQFGELGVLGMHL-DGYGCAGTNAVSYGLACLELEA
TH_2172|M.thermoresistible__bu WFESGQIP-VRELAADLGKLGLLGMHL-QGYGCSGSTATAYGLVCQELEA
MLBr_00737|M.leprae_Br4923 VDQRARFP--EEALAALNASGFNAIHVPEEYGGQGADSVAACIVIEEVAR
: . :* : :. *. .::: . :* *: :. :. *:
Mb0406c|M.bovis_AF2122/97 ADSGIRSLVSVQGSLAMFAIASFGSDEQKRQWLPGMATGDLLGCFGLTEP
Rv0400c|M.tuberculosis_H37Rv ADSGIRSLVSVQGSLAMFAIASFGSDEQKRQWLPGMATGDLLGCFGLTEP
MMAR_0698|M.marinum_M ADSGIRSLVSVQGSLAMFAIAHFGSEEHKQRWLPPMAAGELIGCFGLTEP
MUL_2825|M.ulcerans_Agy99 ADSGIRSLVSVQGSLAMFAIAHFGSEEHKQRWLPPMAAGELIGCFGLTEP
MAV_4767|M.avium_104 ADSGLRSMVSVQGSLAMFAIWNFGSEEQKRQWLPGMATGEMLGCFGLTEP
Mflv_4513|M.gilvum_PYR-GCK GDSGFRSFVSVQGSLSMFSIHRYGSEEQKNEWLPRLASGDAIGCFGLTEP
Mvan_1857|M.vanbaalenii_PYR-1 GDSGFRSFVSVQGSLSMYSIYRYGSDEQKNEWLPQLASGEAIGCFGLTEP
MSMEG_6686|M.smegmatis_MC2_155 GDSGFRSFVSVQGSLSMFSIYRYGSEEQKNEWLPRLAAGDAIGCFGLTEP
MAB_0822|M.abscessus_ATCC_1997 GDSGLRSFVSVQGSLSMYSIHRYGSEEQKQEWLPQLASGHAIGCFGLTEP
TH_2172|M.thermoresistible__bu VDSGLRSLVSVQGSLAMFAIYRHGSEEQRRHWLPPMATGEVIGCFGLTEP
MLBr_00737|M.leprae_Br4923 VDASASLIPAVN-KLGTMGLILRGSEELKKQVLPSLAAEGAMASYALSER
*:. : :*: .*. .: **:* :.. ** :*: :..:.*:*
Mb0406c|M.bovis_AF2122/97 DVGSDPAAMKTRARRDGP----DWVITGGKMWITNGSVADVAIVWAA---
Rv0400c|M.tuberculosis_H37Rv DVGSDPAAMKTRARRDGP----DWVITGGKMWITNGSVADVAIVWAA---
MMAR_0698|M.marinum_M DVGSDPAAMKTRARRDGS----DWVLNGRKMWITNGSIADVAIVWAA---
MUL_2825|M.ulcerans_Agy99 DVGSDPAAMKTRARRDGS----DWVLNGRKMWITNGSIADVAIVWAA---
MAV_4767|M.avium_104 DVGSDPAAMKTRARRDGS----DWVLDGRKLWITNGSVADVAVVWAG---
Mflv_4513|M.gilvum_PYR-GCK DFGSNPAGMRTRGKRDGSGENADWILNGTKMWITNGNLADVATVWAQ---
Mvan_1857|M.vanbaalenii_PYR-1 DFGSNPAGMRTRARRSGAGENADWVLNGTKMWITNGNLADVATVWAQ---
MSMEG_6686|M.smegmatis_MC2_155 DFGSNPAGMRTRARRDGS----DWILNGTKMWITNGNLADVATVWAQ---
MAB_0822|M.abscessus_ATCC_1997 DFGSNPGGMRTRARRDGK----DWILDGTKMWITNGNLADVATVWAQ---
TH_2172|M.thermoresistible__bu DFGSNPAGMRTTARRDGS----DWILNGSKMWITNGSVADVAIVWARS--
MLBr_00737|M.leprae_Br4923 EAGSDAASMRTRAKADGD----DWILNGFKCWITNGGKSTWYTVMAVTDP
: **:...*:* .: .* **:: * * *****. : * *
Mb0406c|M.bovis_AF2122/97 --TDDGIRGFIVPTDTPGFTANTIGHKLSLRASITSELVLDNVRLPADAM
Rv0400c|M.tuberculosis_H37Rv --TDDGIRGFIVPTDTPGFTANTIGHKLSLRASITSELVLDNVRLPADAM
MMAR_0698|M.marinum_M --TEEGIRGFVVPTETAGFAANTIHHKLSLRASITSELVLDDVRLPAEAM
MUL_2825|M.ulcerans_Agy99 --TEEGIRGFVVPTETAGFAANTIHHKLSLRASITSELVLDDVRLPAEAM
MAV_4767|M.avium_104 --TDDGIRGFLVPTDTPGFTANTIHHKLSLRASITSELVLDDVRLPGEAM
Mflv_4513|M.gilvum_PYR-GCK --TDEGIRGFLVPTDTPGFTANAIHRKLSLRASITSELVLDNVRLPASAQ
Mvan_1857|M.vanbaalenii_PYR-1 --TDEGIRGFLVPTDTPGFTANAIHRKLSLRASVTSELVLDNVRLPASAQ
MSMEG_6686|M.smegmatis_MC2_155 --TDDGIRGFLVPTDTPGFTANEIHRKLSLRASVTSELVLDNVRLPASAQ
MAB_0822|M.abscessus_ATCC_1997 --TDDGVRGFVVPTATKGFAANVIHKKLSLRASVTSELVLDGVRLPDSAE
TH_2172|M.thermoresistible__bu --ADDGILGFVVPTDTPGFTAREMTRKLSLRASVTSELHLDDVRLPADAR
MLBr_00737|M.leprae_Br4923 DKGANGISAFIVHKDDEGFSIGPKEKKLGIKGSPTTELYFDKCRIPGDRI
:*: .*:* . **: :**.::.* *:** :* *:* .
Mb0406c|M.bovis_AF2122/97 LPG-ATGLRAPLACLSEARYGIVWGAMGAARSAWQCALDYARQRTQFGRP
Rv0400c|M.tuberculosis_H37Rv LPG-ATGLRAPLACLSEARYGIVWGAMGAARSAWQCALDYARQRTQFGRP
MMAR_0698|M.marinum_M LPG-ATGVRGPLTCLSEARYGIVWGAMGAARSAWQCALDYAKERTQFGRP
MUL_2825|M.ulcerans_Agy99 LPG-ATGVRGPLTCLSEARYGIVWGAMGAARSAWQCALDYAKERTQFGRP
MAV_4767|M.avium_104 LPG-ATGLRGPLSCLSEARYGIIWGAMGAARSAWQAALDYATQRTQFGRP
Mflv_4513|M.gilvum_PYR-GCK LPE-AVGLSAPLSCLNEARFGIVFGALGAARDSLETTIAYTQTREVFDRP
Mvan_1857|M.vanbaalenii_PYR-1 LPD-AVGLSAPLSCLNEARFGIVFGALGAARDSLETTIAYTQTRDVFDKP
MSMEG_6686|M.smegmatis_MC2_155 LPL-AEGLSAPLSCLNEARFGIVFGALGAARDSLETTIAYTQSREVFDKP
MAB_0822|M.abscessus_ATCC_1997 LPG-ARGLSAPLSCLNEARFGIVFGALGAARDSLQAALDYTRTREVFGRP
TH_2172|M.thermoresistible__bu LPG-ARGLSGPLACLSEARFGIVFGSVGAARDCLETTLDYVATREVFDKP
MLBr_00737|M.leprae_Br4923 IGEPGTGFKTALATLDHTRPTIGAQAVGIAQGALDAAIVYTKDRKQFGES
: . *. .*: *..:* * ::* *:.. : :: *. * *...
Mb0406c|M.bovis_AF2122/97 IAGFQLTQAKLVDMAVELHKGQLLSLHLGRLKDRVGLRPDQVSFG-KLNN
Rv0400c|M.tuberculosis_H37Rv IAGFQLTQAKLVDMAVELHKGQLLSLHLGRLKDRVGLRPDQVSFG-KLNN
MMAR_0698|M.marinum_M IAGFQLTQAKLVDMAVELHKGQLLALQLGRLKDSVGLRPEQVSFG-KLNN
MUL_2825|M.ulcerans_Agy99 IAGFQLTQAKLVDMAVELHKGQLLALQLGRLKDSVGLRPEQVSFG-KLNN
MAV_4767|M.avium_104 IAGFQLTQAKLVDMAVELHKGQLLSLHLGRLKDSTGLRPEQVSFG-KLNN
Mflv_4513|M.gilvum_PYR-GCK LAGYQLTQEKLANMTVELGKGMLLAIHLGRIKDAEGVRPEQVSLG-KLNN
Mvan_1857|M.vanbaalenii_PYR-1 LAGYQLTQEKLANMTVELGKGMLLAIHLGRIKDAEGVRPEQISLG-KLNN
MSMEG_6686|M.smegmatis_MC2_155 LSNYQLTQEKLANMTVELGKGMLLAIHLGRIKDAEGVRPEQISLG-KLNN
MAB_0822|M.abscessus_ATCC_1997 LSSYQLSQEKLANMTVQLGMGMLLALHLGRIKDSVGLRPEQVSLG-KLNN
TH_2172|M.thermoresistible__bu LAGYQITQTKLADMAVELGKAQLLALHLGRLKDGGTIRPEQISVG-KLNN
MLBr_00737|M.leprae_Br4923 ISTFQSIQFMLADMAMKVEAARLIVYAAAARAERGEPDLGFISAASKCFA
:: :* * *.:*:::: . *: . : :* . *
Mb0406c|M.bovis_AF2122/97 TREALKICRTARTILGGNGISLEYPVIRHMVNLESVLTYEGTPEMHQLVL
Rv0400c|M.tuberculosis_H37Rv TREALKICRTARTILGGNGISLEYPVIRHMVNLESVLTYEGTPEMHQLVL
MMAR_0698|M.marinum_M TREAIKICRTARTILGGNGISLEYPVIRHMVNLESVLTYEGTPEMHQLVL
MUL_2825|M.ulcerans_Agy99 TREAIKICRTARTILGGNGISLEYPVIRHMVNLESVLTYEGTPEMHQLVL
MAV_4767|M.avium_104 TREAIEICRTARTILGGNGISLEYPVIRHMVNLESVLTYEGTPEMHQLVL
Mflv_4513|M.gilvum_PYR-GCK VREAIAIARECRTLLGGSGITLEYSPLRHANNLESVLTYEGTSEMHLLSI
Mvan_1857|M.vanbaalenii_PYR-1 VREAIAIARECRTLLGGSGITLEYSPLRHANNLESVLTYEGTSEMHLLSI
MSMEG_6686|M.smegmatis_MC2_155 VREAIAIARECRTLLGGSGITLEYSPLRHANNLESVLTYEGTSEMHLLSI
MAB_0822|M.abscessus_ATCC_1997 VREALNIARECRTLLGASGITLEYSPLRHANNLESVLTYEGTSEMHLLSI
TH_2172|M.thermoresistible__bu VREALRIARECRTLLGANGVTLEYPVLRHANNLESVLTYEGTSEVHQLVI
MLBr_00737|M.leprae_Br4923 SDIAMEVTTDAVQLFGGAGYTSDFPVERFMRDAKITQIYEGTNQIQRVVM
*: : . ::*. * : ::. *. : : . **** ::: : :
Mb0406c|M.bovis_AF2122/97 GQAFTGLAAFR-
Rv0400c|M.tuberculosis_H37Rv GQAFTGLAAFR-
MMAR_0698|M.marinum_M GQAFTGLAAFR-
MUL_2825|M.ulcerans_Agy99 GQAFTGLAAFR-
MAV_4767|M.avium_104 GQAFTGADAFR-
Mflv_4513|M.gilvum_PYR-GCK GKALTGHAAFRA
Mvan_1857|M.vanbaalenii_PYR-1 GKALTGHAAFRS
MSMEG_6686|M.smegmatis_MC2_155 GKALTGKAAFRS
MAB_0822|M.abscessus_ATCC_1997 GRALTGQAAFR-
TH_2172|M.thermoresistible__bu GQTMTGLSAFR-
MLBr_00737|M.leprae_Br4923 SRALLR------
.:::