For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TFSTAELPDEYESLRKTVEEFAHSVVAPAAAHHDATKTFPYAVVAQMGEMGLFGLPFPEEYGGMGGDYFA LCLTLEELARADQSVAITLEAAVSLGAMPVYRYGSEEQKQRWLPELASGKSLAAFGLTEPGGGTDIPGSI RTRAVEDNGSWVINGAKAFITNSGTDITAVVGVAAVTGRSADGSPEISVIHVPSGTPGFTVEPGYDKVGW HASDTHPLSFDDVRVPYENLLGERGRGYAGFLRILDEGRIAIAAVATGAAQGCVDESLRYARSRPAFGSA IIDYQSVSFKIARMQARAHTARLAYYHAAALLLAGKPFKTEASIAKLIASEAAMDNARDATQIFGGAGFM NESPVARQYRDSKILEIGEGTSEVQLMLIARALTG*
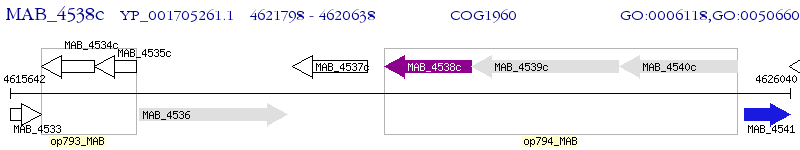
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4538c | - | - | 100% (386) | putative acyl-CoA dehydrogenase FadE |
| M. abscessus ATCC 19977 | MAB_3618c | - | 5e-65 | 37.50% (384) | acyl-CoA dehydrogenase FadE |
| M. abscessus ATCC 19977 | MAB_1863 | - | 2e-57 | 35.54% (377) | acyl-CoA dehydrogenase FadE |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2528c | fadE19 | 1e-157 | 72.02% (386) | acyl-CoA dehydrogenase |
| M. gilvum PYR-GCK | Mflv_2554 | - | 1e-159 | 74.28% (381) | acyl-CoA dehydrogenase domain-containing protein |
| M. tuberculosis H37Rv | Rv2500c | fadE19 | 1e-157 | 72.02% (386) | acyl-CoA dehydrogenase |
| M. leprae Br4923 | MLBr_00737 | fadE25 | 7e-68 | 38.70% (385) | putative acyl-CoA dehydrogenase |
| M. marinum M | MMAR_3847 | fadE19 | 1e-160 | 73.23% (381) | acyl-CoA dehydrogenase FadE19 |
| M. avium 104 | MAV_1672 | - | 1e-158 | 72.85% (383) | putative acyl-CoA dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_4715 | - | 1e-162 | 75.33% (381) | acyl-CoA dehydrogenase |
| M. thermoresistible (build 8) | TH_2782 | - | 1e-162 | 75.59% (381) | PUTATIVE acyl-CoA dehydrogenase domain protein |
| M. ulcerans Agy99 | MUL_3778 | fadE19 | 1e-159 | 72.70% (381) | acyl-CoA dehydrogenase FadE19 |
| M. vanbaalenii PYR-1 | Mvan_4089 | - | 1e-160 | 75.07% (381) | acyl-CoA dehydrogenase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2554|M.gilvum_PYR-GCK -----MSDFLSTGTLPDHYAQLAKTVRDFAQSVVAPVAAKHDEEHSFPYE
MSMEG_4715|M.smegmatis_MC2_155 -----MTDFLSTGSLPDDYEQLAKTVRDFAQSVVAPVAAKHDEEHSFPYE
Mvan_4089|M.vanbaalenii_PYR-1 -----MSNNLATGILPDHYEQLAKTVRDFAQSVVAPVAAKHDEEHSFPYE
TH_2782|M.thermoresistible__bu -----MPDYLTTGNLPDHYAELAGTVRDFAQSVVAPVAAKHDQERSFPYE
Mb2528c|M.bovis_AF2122/97 --MTTTTTTISGGILPKEYQDLRDTVADFARTVVAPVSAKHDAEHSFPYE
Rv2500c|M.tuberculosis_H37Rv --MTTTTTTISGGILPKEYQDLRDTVADFARTVVAPVSAKHDAEHSFPYE
MMAR_3847|M.marinum_M -----MTTAISAGTLPKEYQDLRDTVADFARSVVAPVSAKHDEEHSFPYE
MUL_3778|M.ulcerans_Agy99 -----MTTAISAGTLPKEYQDLRDTVADFARSVVAPVSAKHDEEHSFPYE
MAV_1672|M.avium_104 -----MTTSITAGTLPKEYQDLRDTVAEFARTVVAPVSAKHDEEHSFPYE
MAB_4538c|M.abscessus_ATCC_199 -------MTFSTAELPDEYESLRKTVEEFAHSVVAPAAAHHDATKTFPYA
MLBr_00737|M.leprae_Br4923 MVGWSGNPLFDLFKLPEEHNELRATIRALAEKEIAPHAADVDQRARFPEE
: **..: .* *: :*.. :** :*. * **
Mflv_2554|M.gilvum_PYR-GCK VVSGMADMGLFGLPFPEEYGGMGGDYFALCLALEELGKVDQSVAITLEAG
MSMEG_4715|M.smegmatis_MC2_155 VVAGMADMGLFGLPFPEEYGGMGGDYFALCLALEELGRVDQSVAITLEAG
Mvan_4089|M.vanbaalenii_PYR-1 VVAGMADMGLFGLPFPEEYGGMGGDYFALCLALEELGKVDQSVAITLEAG
TH_2782|M.thermoresistible__bu VVAAMGEMGLFGLPFPEEYGGMGGDYFALCLALEELGKVDQSVAITLEAG
Mb2528c|M.bovis_AF2122/97 IVAKMGEMGLFGLPFPEEYGGMGGDYFALSLVLEELGKVDQSVAITLEAA
Rv2500c|M.tuberculosis_H37Rv IVAKMGEMGLFGLPFPEEYGGMGGDYFALSLVLEELGKVDQSVAITLEAA
MMAR_3847|M.marinum_M VVAKMGEMGLFGLPFPEEYGGMGGDYFALALALEELGKVDQSVAITLEAG
MUL_3778|M.ulcerans_Agy99 VVAKMGEMGLFGLPFPEEYGGMGGDYFALALALEELGKVNQSVAITLEAG
MAV_1672|M.avium_104 VVAKMGEMGLFGLPFPEEYGGMGGDYFALSLALEELGKVDQSVAITLEAG
MAB_4538c|M.abscessus_ATCC_199 VVAQMGEMGLFGLPFPEEYGGMGGDYFALCLTLEELARADQSVAITLEAA
MLBr_00737|M.leprae_Br4923 ALAALNASGFNAIHVPEEYGGQGADSVAACIVIEEVARVDASASL-IPAV
:: : *: .: .****** *.* .* .:.:**:.:.: *.:: : *
Mflv_2554|M.gilvum_PYR-GCK VSLGAMPVYRFGNESQKQEWLPLLASGKALGAFGLTEAGGGSD-AGATKT
MSMEG_4715|M.smegmatis_MC2_155 VSLGAMPVYRFGNDAQKQEWLPLLASGKALGAFGLTEAGGGSD-AGATKT
Mvan_4089|M.vanbaalenii_PYR-1 VSLGAMPVYRFGTEAQKQEWLPLLASGKALGAFGLTEAGGGSD-AGATKT
TH_2782|M.thermoresistible__bu VSLGAMPIYRFGTEEQKQRWLPELAAGKALGAFGLTEPGGGSD-AGATRT
Mb2528c|M.bovis_AF2122/97 VGLGAMPIYRFGTEEQKQKWLPDLTSGRALAGFGLTEPGAGSD-AGSTRT
Rv2500c|M.tuberculosis_H37Rv VGLGAMPIYRFGTEEQKQKWLPDLTSGRALAGFGLTEPGAGSD-AGSTRT
MMAR_3847|M.marinum_M VGLGAMPIYRFGNEEQKSKWLPDLLAGRALAGFGLTEPGAGSD-AGSTRT
MUL_3778|M.ulcerans_Agy99 VGLGAMPIYRFGNEEQKSKWLPDLLAGRALAGFGLTEPGAGSD-AGSTRT
MAV_1672|M.avium_104 VSLGAMPIYRFGNEEQKRKWLPDLTAGRALAGFGLTEPGAGSD-AGATRT
MAB_4538c|M.abscessus_ATCC_199 VSLGAMPVYRYGSEEQKQRWLPELASGKSLAAFGLTEPGGGTDIPGSIRT
MLBr_00737|M.leprae_Br4923 NKLGTMGLILRGSEELKKQVLPSLAAEGAMASYALSEREAGSD-AASMRT
**:* : *.: * . ** * : ::..:.*:* .*:* ..: :*
Mflv_2554|M.gilvum_PYR-GCK TAKSDGASWVINGTKQFITNSGTDITKLVTVTAVTG---EN-NGKKEISS
MSMEG_4715|M.smegmatis_MC2_155 TARLDDGHWVINGSKQFITNSGTDITRLVTVTAVTG---EKPDGRKEISS
Mvan_4089|M.vanbaalenii_PYR-1 TARLDDGHWVINGSKQFITNSGTDITKLVTVTAVTG---ET-DGKKEISS
TH_2782|M.thermoresistible__bu TARLDDGHWVINGSKQFITNSGTDITKLVTVTAVTG---EV-AGKKEISA
Mb2528c|M.bovis_AF2122/97 TARLEGDEWIINGSKQFITNSGTDITSLVTVTAVTGTTGTAADAKKEIST
Rv2500c|M.tuberculosis_H37Rv TARLEGDEWIINGSKQFITNSGTDITSLVTVTAVTGTTGTAADAKKEIST
MMAR_3847|M.marinum_M TARLDGGEWVVNGSKQFITNSGTDITSLVTITAVTG---SIADGKKEIST
MUL_3778|M.ulcerans_Agy99 TARLDGGEWVVNGSKQFITNSGTDITSLVTITAVTG---SIADGKKEIST
MAV_1672|M.avium_104 TARLEDGEWVINGSKQFITNSGTDITSLVTVTAVTG---TVGADKKEIST
MAB_4538c|M.abscessus_ATCC_199 RAVEDNGSWVINGAKAFITNSGTDITAVVGVAAVTG---RSADGSPEISV
MLBr_00737|M.leprae_Br4923 RAKADGDDWILNGFKCWITNGGK--STWYTVMAVTD----PDKGANGISA
* :. *::** * :***.*. : : ***. **
Mflv_2554|M.gilvum_PYR-GCK ILVPVPTEGFTAEPAYNKVGWNASDTHPLSFDDVRVPQENLLGERGRGYA
MSMEG_4715|M.smegmatis_MC2_155 ILVPVPTEGFTAEPAYNKVGWNASDTHPLSFDDVRVPEENLLGERGRGYA
Mvan_4089|M.vanbaalenii_PYR-1 ILVPVPTPGFTAEPAYNKVGWNASDTHPLSFDDVRVPQENLLGERGRGYA
TH_2782|M.thermoresistible__bu IIVPVPTDGFTVEPAYHKVGWNASDTHPLSFDDVRVPEENLLGERGRGYA
Mb2528c|M.bovis_AF2122/97 IIVPSGTPGFTVEPVYNKVGWNASDTHPLTFADARVPRENLLGARGSGYA
Rv2500c|M.tuberculosis_H37Rv IIVPSGTPGFTVEPVYNKVGWNASDTHPLTFADARVPRENLLGARGSGYA
MMAR_3847|M.marinum_M IIVPSGTPGFIVEPVYNKVGWNASDTHPLSFDDARVPEENLLGIRGKGYA
MUL_3778|M.ulcerans_Agy99 IIVPSGTPGFIVEPVYNKVGWNASDTHPLSFDDARVPEENLLGIRGKGYA
MAV_1672|M.avium_104 IIVPSGTPGFTVEPVYSKVGWNASDTHPLSFSDARVPEENLLGARGTGYA
MAB_4538c|M.abscessus_ATCC_199 IHVPSGTPGFTVEPGYDKVGWHASDTHPLSFDDVRVPYENLLGERGRGYA
MLBr_00737|M.leprae_Br4923 FIVHKDDEGFSIGPKEKKLGIKGSPTTELYFDKCRIPGDRIIGEPGTGFK
: * ** * *:* :.* * * * . *:* :.::* * *:
Mflv_2554|M.gilvum_PYR-GCK NFLRILDEGRIAIAALSVGAAQGCVDESVKYAKERQAFGAAIGTYQAIAF
MSMEG_4715|M.smegmatis_MC2_155 NFLRILDEGRIAIAALSVGAAQGCVDESVKYAKERQAFGAAIGTYQAIAF
Mvan_4089|M.vanbaalenii_PYR-1 GFLRILDEGRIAIAALSVGAAQGCVDESVKYAKERHAFGRAIGTNQAIAF
TH_2782|M.thermoresistible__bu NFLRILDEGRIAIAALSVGAAQGCVDESVKYAKEREAFGRPIGANQAIAF
Mb2528c|M.bovis_AF2122/97 NFLSILDEGRIAIAALATGAAQGCVDESVKYANQRQSFGQPIGAYQAIGF
Rv2500c|M.tuberculosis_H37Rv NFLSILDEGRIAIAALATGAAQGCVDESVKYANQRQSFGQPIGAYQAIGF
MMAR_3847|M.marinum_M NFLSILDEGRIAIAALATGVAQGCVDESVKYAKERQSFGQPIGSYQAISF
MUL_3778|M.ulcerans_Agy99 NFLSILDEGRIAIAALATGVAQGCVDESVKYAKERQSFGQPIGSYQAISF
MAV_1672|M.avium_104 NFLSILDEGRIAIAALATGAAQGCVDESVKYAKERQSFGQPIGSYQAISF
MAB_4538c|M.abscessus_ATCC_199 GFLRILDEGRIAIAAVATGAAQGCVDESLRYARSRPAFGSAIIDYQSVSF
MLBr_00737|M.leprae_Br4923 TALATLDHTRPTIGAQAVGIAQGALDAAIVYTKDRKQFGESISTFQSIQF
* **. * :*.* :.* ***.:* :: *:..* ** .* *:: *
Mflv_2554|M.gilvum_PYR-GCK KIARMEARAHTARAAYYDAAALMLSGKP-FKKAASVAKMVASEAAMDNAR
MSMEG_4715|M.smegmatis_MC2_155 KIARMEARAHAARTAYYDAAALMLAGKP-FKKAASVAKLVASEAAMDNAR
Mvan_4089|M.vanbaalenii_PYR-1 KIARMEARAHAARAAYYDAAALMLAGKP-FKKAAAVAKLVSSEAAMDNAR
TH_2782|M.thermoresistible__bu KIARMEARAHTARTAYYDAAALMLAGKP-FKKQAAIAKMVASEAAMDNAR
Mb2528c|M.bovis_AF2122/97 KIARMEARAHVARTAYYDAAAKMLAGKP-FKKEAAIAKMISSEAAMDNSR
Rv2500c|M.tuberculosis_H37Rv KIARMEARAHVARTAYYDAAAKMLAGKP-FKKEAAIAKMISSEAAMDNSR
MMAR_3847|M.marinum_M KIARMEARAHVARTAYYEAAAKMLAGKP-FKKEAAIAKMISSEAAMDNAR
MUL_3778|M.ulcerans_Agy99 KIARMEARAHVARTAYYEAAAKMLAGKP-FKKEAAIAKMISSEAAMDNAR
MAV_1672|M.avium_104 KIARMEARAHVARTAYYDAAAKMLAGKP-FKKEAAIAKMIASEAAMDNAR
MAB_4538c|M.abscessus_ATCC_199 KIARMQARAHTARLAYYHAAALLLAGKP-FKTEASIAKLIASEAAMDNAR
MLBr_00737|M.leprae_Br4923 MLADMAMKVEAARLIVYAAAARAERGEPDLGFISAASKCFASDIAMEVTT
:* * :...** * *** *:* : :: :* .:*: **: :
Mflv_2554|M.gilvum_PYR-GCK DATQIFGGYGFMNEYSVARHYRDSKILEIGEGTTEVQLMLIGRELGL-
MSMEG_4715|M.smegmatis_MC2_155 DATQIFGGYGFMNEYSVARHYRDSKILEIGEGTTEVQLMLIGRELGL-
Mvan_4089|M.vanbaalenii_PYR-1 DATQIFGGYGFMNEYPVARHYRDSKILEIGEGTTEVQLMLIGRELGL-
TH_2782|M.thermoresistible__bu DATQIFGGYGFINEFPVARHYRDSKILEIGEGTTEVQLMLIGRELGL-
Mb2528c|M.bovis_AF2122/97 DATQIHGGYGFMNEYPVARHYRDSKVLEIGEGTTEVQLMLIARSLGLQ
Rv2500c|M.tuberculosis_H37Rv DATQIHGGYGFMNEYPVARHYRDSKVLEIGEGTTEVQLMLIARSLGLQ
MMAR_3847|M.marinum_M DATQVHGGYGFMNEYPVARHYRDSKILEIGEGTTEVQLMLIARSLGLS
MUL_3778|M.ulcerans_Agy99 DAAQVHGGYGFMNEYPVARHYRDSKILEIGEGTTEVQLMLIARSLGLS
MAV_1672|M.avium_104 DATQIHGGYGFMNEYPVARHYRDSKILEIGEGTTEVQLMLIARSLGLS
MAB_4538c|M.abscessus_ATCC_199 DATQIFGGAGFMNESPVARQYRDSKILEIGEGTSEVQLMLIARALTG-
MLBr_00737|M.leprae_Br4923 DAVQLFGGAGYTSDFPVERFMRDAKITQIYEGTNQIQRVVMSRALLR-
**.*:.** *: .: .* * **:*: :* ***.::* :::.* *