For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTSVYSLQGGPAVAQVRRMATEVPGPRSRELAERRAAALPAGLVSGAGVYAAAAGGGVLVDVDGNSFLDL GSGIAVTTVGNSAPAVVARVAAQAQRFTHTCFLATPYEQYLEVAETLNRITPGDHEKRTALFNTGAEAVE NAVKYARVATARPAVVVFDHAFHGRSLMTMTMTAKQAPYRRGFGPFAPEVYRAPMAHPYRWPSGAENCAS DAFRQFASLVDNQIGAEAVACVVVEPIQGEGGFIVPAPGFLAMVADFCRARGILLVADEVQTGIARTGAW FASEHEQLVPDLVTTAKGLAGGLPLAAVTGRADVMDAAHPGGIGGTFSGNPLSCAAALGVFEEIETHDLL RRAADMGEMLLREMRGISAETGLIGDIRGRGAMIAAELVVPGTDSPNREAVIQILKYCQDNGVLVLSAGT YGNVLRFLPPLSMPDELLQEALAVLRDAFRNL
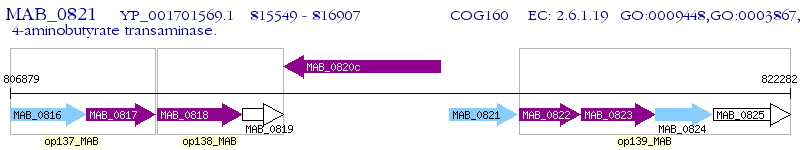
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0821 | - | - | 100% (452) | 4-aminobutyrate aminotransferase GabT |
| M. abscessus ATCC 19977 | MAB_4207 | - | e-143 | 56.62% (438) | 4-aminobutyrate aminotransferase |
| M. abscessus ATCC 19977 | MAB_4417c | - | 2e-66 | 35.93% (398) | aminotransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2620 | gabT | 1e-134 | 53.86% (440) | 4-aminobutyrate aminotransferase |
| M. gilvum PYR-GCK | Mflv_3818 | - | 1e-144 | 57.59% (448) | 4-aminobutyrate aminotransferase |
| M. tuberculosis H37Rv | Rv2589 | gabT | 1e-134 | 53.86% (440) | 4-aminobutyrate aminotransferase |
| M. leprae Br4923 | MLBr_00485 | gabT | 1e-137 | 54.23% (437) | 4-aminobutyrate aminotransferase |
| M. marinum M | MMAR_2118 | gabT | 1e-134 | 54.15% (434) | 4-aminobutyrate aminotransferase GabT |
| M. avium 104 | MAV_3470 | gabT | 1e-139 | 56.06% (437) | 4-aminobutyrate aminotransferase |
| M. smegmatis MC2 155 | MSMEG_6685 | gabT | 0.0 | 74.50% (447) | 4-aminobutyrate transaminase |
| M. thermoresistible (build 8) | TH_3529 | - | 2e-50 | 35.71% (406) | PUTATIVE aminotransferase |
| M. ulcerans Agy99 | MUL_1723 | gabT | 1e-132 | 53.46% (434) | 4-aminobutyrate aminotransferase |
| M. vanbaalenii PYR-1 | Mvan_1858 | - | 0.0 | 76.96% (447) | 4-aminobutyrate aminotransferase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_6685|M.smegmatis_MC2_155 MT--ATLTGGPDLQQARRILTEIPGPRSRDLAERRNAALPAGLASGAGVY
Mvan_1858|M.vanbaalenii_PYR-1 MT--TSLVGGPALPQARRMVTDIPGPRSAELAARRSAALPAGLASAAGVY
MAB_0821|M.abscessus_ATCC_1997 MTSVYSLQGGPAVAQVRRMATEVPGPRSRELAERRAAALPAGLVSGAGVY
MMAR_2118|M.marinum_M MA---------SLDQSRLLVTEIPGPASLELNKRRAAAVSGGVGVTLPVF
MUL_1723|M.ulcerans_Agy99 MA---------SLDQSRLLVTEIPGPASLELNKRRAAAVSGGVGVTLPVF
MAV_3470|M.avium_104 MA---------RLEQTRRLVTEIPGPASLELSKRRAAAVSHAVNVSVPVF
Mb2620|M.bovis_AF2122/97 MA---------SLQQSRRLVTEIPGPASQALTHRRAAAVSSGVGVTLPVF
Rv2589|M.tuberculosis_H37Rv MA---------SLQQSRRLVTEIPGPASQALTHRRAAAVSSGVGVTLPVF
MLBr_00485|M.leprae_Br4923 MT---------SVEQSRQLVTEIPGPVSLELAKRLNAAVPRGVGVTLPVF
Mflv_3818|M.gilvum_PYR-GCK MS---------ALEQSRHLATAIPGPRSVELASRKNTAVSRGVGTTMPVY
TH_3529|M.thermoresistible__bu ----------------------VSGSKGRHTLLDRWQGVMMNNYGTPPLA
:.*. . .: :
MSMEG_6685|M.smegmatis_MC2_155 IAAAGGGVVVDVDGNSFIDLGSGIAVTTVGNAAPAVVSRAGSQLAQYTHT
Mvan_1858|M.vanbaalenii_PYR-1 VAAAGGGVAVDVDGNSFIDLGSGIAVTTVGNSAPAVVERATAQLARYTHT
MAB_0821|M.abscessus_ATCC_1997 AAAAGGGVLVDVDGNSFLDLGSGIAVTTVGNSAPAVVARVAAQAQRFTHT
MMAR_2118|M.marinum_M VVRAGGGIVEDADGNRLIDLGSGIAVTTIGNSAPRVVDAVRDQVEQFTHT
MUL_1723|M.ulcerans_Agy99 VVRAGGGIVEDADGNRLIDLGSGIAVTTIGNSAPRVVDAVRDQVEQFTHT
MAV_3470|M.avium_104 VARAGGGIVEDVDGNRLIDLGSGIAVTTIGNSAPRVVDAVRAQVAEFTHT
Mb2620|M.bovis_AF2122/97 VARAGGGIVEDVDGNRLIDLGSGIAVTTIGNSSPRVVDAVRTQVAEFTHT
Rv2589|M.tuberculosis_H37Rv VARAGGGIVEDVDGNRLIDLGSGIAVTTIGNSSPRVVDAVRTQVAEFTHT
MLBr_00485|M.leprae_Br4923 VTRAAGGVIEDVDGNRLIDLGSGIAVTTIGNSSPRVVDAVRAQVADFTHT
Mflv_3818|M.gilvum_PYR-GCK AARAFGGIVEDVDGNRLIDLGSGIAVTTVGNASPRVVAAVAEQAAEFTHT
TH_3529|M.thermoresistible__bu LVSGDGAVVTDADGKSYLDLLGGIAVNILGHRHPAVIEAVTRQLNTLGHT
. . *.: *.**: :** .****. :*: * *: . * **
MSMEG_6685|M.smegmatis_MC2_155 CFLNVPYEPYIEVAERLNALTPGDHQKRTALFSTGAEAVENAVKYARAYT
Mvan_1858|M.vanbaalenii_PYR-1 CFLATPYEPYIEVAETLNRLTPGSHDKRTALFNTGSEAVENAVKYARAAT
MAB_0821|M.abscessus_ATCC_1997 CFLATPYEQYLEVAETLNRITPGDHEKRTALFNTGAEAVENAVKYARVAT
MMAR_2118|M.marinum_M CFMVTPYEQYVAVAEQLNRITPGSGEKRTVLFNSGAEAVENSIKVARAHT
MUL_1723|M.ulcerans_Agy99 CFMVIPYEHYVAVAEQLNRITPGSGEKRTVLFNSGAEAVENSIKVARAHT
MAV_3470|M.avium_104 CFMVTPYESYVAVAETLNRITPGAGEKRSVLFNSGAEAVENAVKVARAYT
Mb2620|M.bovis_AF2122/97 CFMVTPYEGYVAVAEQLNRITPGSGPKRSVLFNSGAEAVENAVKIARSYT
Rv2589|M.tuberculosis_H37Rv CFMVTPYEGYVAVAEQLNRITPGSGPKRSVLFNSGAEAVENAVKIARSYT
MLBr_00485|M.leprae_Br4923 CFIITPYEEYVAVTEQLNRITPGSGEKRSVLFNSGAEAVENSIKVARSYT
Mflv_3818|M.gilvum_PYR-GCK CFMVTPYEGYVAVAEALNRLTPGDGDKRSALFNSGSEAVENAVKIARTFT
TH_3529|M.thermoresistible__bu SNLYATEPGVALAEALVGHLG---APARVFFCNSGAEANEVAFKISR-LT
. : . . :. : * : .:*:** * :.* :* *
MSMEG_6685|M.smegmatis_MC2_155 GRDAVVVFDHAFHGRSLLTMTMTAKNQPYKHGFGPFAPEVYRAPMAYPYR
Mvan_1858|M.vanbaalenii_PYR-1 GRSAVVVFDHAFHGRSLLTMTMTAKNQPYKHGFGPFAPEVYRAPMAYPYR
MAB_0821|M.abscessus_ATCC_1997 ARPAVVVFDHAFHGRSLMTMTMTAKQAPYRRGFGPFAPEVYRAPMAHPYR
MMAR_2118|M.marinum_M RKQAVVAFDYAYHGRTNLTMALTAKSMPYKSGFGPFAPEIYRAPVSYPYR
MUL_1723|M.ulcerans_Agy99 RKQAVVAFDYAYHGRTNLTMALTAKSMPYKSGFGPFAPEIYRAPVSYPYR
MAV_3470|M.avium_104 RKPAVVAFDHAYHGRTNLAMALTAKSMPYKGGFGPFAPEIYRAPLSYPYR
Mb2620|M.bovis_AF2122/97 GKPAVVAFDHAYHGRTNLTMALTAKSMPYKSGFGPFAPEIYRAPLSYPYR
Rv2589|M.tuberculosis_H37Rv GKPAVVAFDHAYHGRTNLTMALTAKSMPYKSGFGPFAPEIYRAPLSYPYR
MLBr_00485|M.leprae_Br4923 RKPAVVAFDHAYHGRTNLTMALTAKSMPYKSGFGPFAPEIYRAPLSYPYR
Mflv_3818|M.gilvum_PYR-GCK RKQAVVSFDHAYHGRTNLTMALTAKSMPYKHGFGPFAPEIYRAPMSYPFR
TH_3529|M.thermoresistible__bu GRTKVVAAEGGFHGRTMGALALTGQP-SKREPFEPLPGDVIHVPYGDAG-
: ** : .:***: ::::*.: . : * *:. :: :.* . .
MSMEG_6685|M.smegmatis_MC2_155 WPSGP----ENCAAEAFDLFTTLIDAQIGADQVACVVVEPIQGEGGFIVP
Mvan_1858|M.vanbaalenii_PYR-1 WPSGP----QHCAEEAFAHFAQLVDAQIGADAVACVVVEPIQGEGGFIVP
MAB_0821|M.abscessus_ATCC_1997 WPSGA----ENCASDAFRQFASLVDNQIGAEAVACVVVEPIQGEGGFIVP
MMAR_2118|M.marinum_M DNLLD-KDIATDGELAAERAINLIDKQIGAANLAAVIIEPIAGEGGFIVP
MUL_1723|M.ulcerans_Agy99 DNLLD-KDIATDGELAAERAINLIDKQIGAANLAAVIIEPIAGEGGFIVP
MAV_3470|M.avium_104 DGLLD-KELATDGEKAAARAINIIDGQIGADSLAAVIIEPIQGEGGFIVP
Mb2620|M.bovis_AF2122/97 DGLLD-KQLATNGELAAARAIGVIDKQVGANNLAALVIEPIQGEGGFIVP
Rv2589|M.tuberculosis_H37Rv DGLLD-KQLATNGELAAARAIGVIDKQVGANNLAALVIEPIQGEGGFIVP
MLBr_00485|M.leprae_Br4923 DGLLN-KDLATDGKLAGARAINVIEKQVGADDLAAVIIEPIQGEGGFIVP
Mflv_3818|M.gilvum_PYR-GCK DAEFGGKELATDGELAARRAITVIDKQVGAGNLAAVIIEPIQGEGGFIVP
TH_3529|M.thermoresistible__bu -----------------------ALERAVDDTTAAVFLEPIMGEGGVVVP
: *.:.:*** ****.:**
MSMEG_6685|M.smegmatis_MC2_155 AEGFLQKVADFCRERGILVVADEVQTGIARTGAWFACEHED---LVPDLI
Mvan_1858|M.vanbaalenii_PYR-1 ADGFLRAVAEFCRERGILLVADEVQTGIARTGAWFACEHDG---IVPDLI
MAB_0821|M.abscessus_ATCC_1997 APGFLAMVADFCRARGILLVADEVQTGIARTGAWFASEHEQ---LVPDLV
MMAR_2118|M.marinum_M ADGFLPALQRWCRDNDVVFIADEVQTGFARTGAMFACDHEN---VEPDLI
MUL_1723|M.ulcerans_Agy99 ADGFLPALQRWSRDNDVVFIADEVQTGFARTGAMFACDHEN---VEPDLI
MAV_3470|M.avium_104 AEGFLPTLLDWCRRNDVLFIADEVQTGFARSGAMFACEHEGPNGLEPDLI
Mb2620|M.bovis_AF2122/97 AEGFLPALLDWCRKNHVVFIADEVQTGFARTGAMFACEHEGPDGLEPDLI
Rv2589|M.tuberculosis_H37Rv AEGFLPALLDWCRKNHVVFIADEVQTGFARTGAMFACEHEGPDGLEPDLI
MLBr_00485|M.leprae_Br4923 AEGFLATLLDWCRKNNVMFIADEVQTGFARTGAMFACEHDG---IVPDLI
Mflv_3818|M.gilvum_PYR-GCK AEGFLPTLLDWCRANDVVFIADEVQTGFARTGAMFACEHEGPDGLVPDLI
TH_3529|M.thermoresistible__bu PAGYLAAAREVTARHGALLVLDEVQTGIGRTGTFFAHQHDG---ITPDVV
. *:* . :.: ******:.*:*: ** :*: : **::
MSMEG_6685|M.smegmatis_MC2_155 TTAKGLGGGLPLAAVTGRAEIMDAAHAGGIGGTYAGNPVACAAALGVFDE
Mvan_1858|M.vanbaalenii_PYR-1 TTAKGLAGGLPLAAVTGRADVMDAAHPGGIGGTYSGNPVACAAALGVFEE
MAB_0821|M.abscessus_ATCC_1997 TTAKGLAGGLPLAAVTGRADVMDAAHPGGIGGTFSGNPLSCAAALGVFEE
MMAR_2118|M.marinum_M VTAKGIADGFPLSAVTGRAEIMDAPHTSGLGGTFGGNPVACAAALATIET
MUL_1723|M.ulcerans_Agy99 VTAKGIADGFPLSAVTGRAEIMDAPHTSGLGGTFGGNPVACAAALATIET
MAV_3470|M.avium_104 CTAKAIADGLPLSAVTGRAQIMDAPHVGGLGGTFGGNPLACAAALATIET
Mb2620|M.bovis_AF2122/97 CTAKGIADGLPLSAVTGRAEIMNAPHVGGLGGTFGGNPVACAAALATIAT
Rv2589|M.tuberculosis_H37Rv CTAKGIADGLPLSAVTGRAEIMNAPHVGGLGGTFGGNPVACAAALATIAT
MLBr_00485|M.leprae_Br4923 CTAKGIADGLPLAAVTGRAEIMNAPHVSGLGGTFGGNPVACAAALATITT
Mflv_3818|M.gilvum_PYR-GCK VTAKGIAGGMPLSAVTGRADIMDAPHVSGLGGTYGGNPVACAAALATIET
TH_3529|M.thermoresistible__bu TLAKGLGGGLPIGACLAVGPAGDLLTPGMHGSTFGGNPVCTAAAVAVLGA
**.:..*:*:.* . . : . *.*:.***:. ***:..:
MSMEG_6685|M.smegmatis_MC2_155 IEQNGLIERARAIGEVIVAELRSIAATTGLIGEIRGRGAMIAVELIKPGT
Mvan_1858|M.vanbaalenii_PYR-1 IESGRLVERARTIGEAMVTALEDIAAGTDVIGEIRGRGAMIAAELVVPGT
MAB_0821|M.abscessus_ATCC_1997 IETHDLLRRAADMGEMLLREMRGISAETGLIGDIRGRGAMIAAELVVPGT
MMAR_2118|M.marinum_M IERDGMVERARQIERLVMDRLLRLQAADDRLGDVRGRGAMIAMELVKSGT
MUL_1723|M.ulcerans_Agy99 IERDGMVERARQIERLVMDRLLRLQAADDRLGDVRGRGAMIAMELVKSGT
MAV_3470|M.avium_104 IESDGLIERARQLERLITEPLLRLQARDDRIGDVRGRGAMIAIELVKSGT
Mb2620|M.bovis_AF2122/97 IESDGLIERARQIERLVTDRLTTLQAVDDRIGDVRGRGAMIAVELVKSGT
Rv2589|M.tuberculosis_H37Rv IESDGLIERARQIERLVTDRLTTLQAVDDRIGDVRGRGAMIAVELVKSGT
MLBr_00485|M.leprae_Br4923 IENDGLIQRAQQIERLITDRLLRLQDADDRIGDVRGRGAMIAVELVKSGT
Mflv_3818|M.gilvum_PYR-GCK IESDNLVARAAQIEALMKDRLGRMQAEDDRIGDVRGRGAMIAVELVKSGT
TH_3529|M.thermoresistible__bu LAEGDLINRADVLGKKLSHGIEELRHP--LVDHVRGRGLLCGVVLNAP--
: :: ** : : : : :..:**** : . * .
MSMEG_6685|M.smegmatis_MC2_155 REPNKEAVAALAKHCHDNGVLTLTAGTFGNVLRFLPPLTITDELLVDAFG
Mvan_1858|M.vanbaalenii_PYR-1 REPNRDAVAAISRHCHLNGVLTLTAGTFGNVMRFLPPLSISDELLTEAFD
MAB_0821|M.abscessus_ATCC_1997 DSPNREAVIQILKYCQDNGVLVLSAGTYGNVLRFLPPLSMPDELLQEALA
MMAR_2118|M.marinum_M AEPDAALTQKLAAAAHAAGVIVLTCGMFGNVIRLLPPLTISDELLSEGLD
MUL_1723|M.ulcerans_Agy99 AEPDAALTQKLAAAAHAAGVIVLTCGMFGNVIRLLPPLTISDELLSEGLD
MAV_3470|M.avium_104 AEPDKDLTQRLSVAAHAAGVIVLTCGMFGNIIRLLPPLTISDELLAEGID
Mb2620|M.bovis_AF2122/97 TEPDAGLTERLATAAHAAGVIILTCGMFGNIIRLLPPLTIGDELLSEGLD
Rv2589|M.tuberculosis_H37Rv TEPDAGLTERLATAAHAAGVIILTCGMFGNIIRLLPPLTIGDELLSEGLD
MLBr_00485|M.leprae_Br4923 AEPDPELTEKVATAAHATGVIILTCGMFGNIIRLLPPLTISDELLAEGLD
Mflv_3818|M.gilvum_PYR-GCK LEPDPELTRKLLTAAHAAGVIVLSCGTFGNVLRFLPPLAISDDLLLEGLD
TH_3529|M.thermoresistible__bu --------QAKAAEAAARDAGFLVNAAAPEVIRLAPPLIITETQIDEFLA
. .. * . :::*: *** : : : : :
MSMEG_6685|M.smegmatis_MC2_155 VIRDGFAAL----
Mvan_1858|M.vanbaalenii_PYR-1 VVRDGFASL----
MAB_0821|M.abscessus_ATCC_1997 VLRDAFRNL----
MMAR_2118|M.marinum_M ILCQILADL----
MUL_1723|M.ulcerans_Agy99 ILCQILADL----
MAV_3470|M.avium_104 ILGGLLADL----
Mb2620|M.bovis_AF2122/97 IVCAILADL----
Rv2589|M.tuberculosis_H37Rv IVCAILADL----
MLBr_00485|M.leprae_Br4923 ILSRILGDF----
Mflv_3818|M.gilvum_PYR-GCK ILELILRDL----
TH_3529|M.thermoresistible__bu ALPGVLDTAKEAS
: :