For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTTSTNTAPSGSGRALNPTLRRVDVFAVLVCALVGVDTLGAVSAYGPQGLLWMALVAVIFFLPYGLLTAE LGSAFPQEGGPYVWTKLAFGKFAAGINQFFYWISNPVWIGGTLCIVAVVTFEEFFFPLPGLWKWVAAAVF VWGSACVIGASVRFGRWIFLVGAAARIALLGLFIASVIVYAYKNGLQPMHTSDFSWTYAGFVAVIPVIVF NFLGFEVPSNAAEEMVNPQKDVPLTVLRGGLTATLLYGIPVLGILLVLPKNQLGTVTGFVDACRAVFTVY GGSVAPDGTAELHGLGQAAAQVAAAGLIIGLFTSGTAWAMGVSRAQAVAFTDGAGPAWLGKFSGNGSPAR VNIMSAVVSTIVMAAAMIFAHGNAEKYFSAAIGLVVSATAISYILTFSSFIRLRHKYPHTRRPFQVFGGR IGGWIVTVLTVGTMLFTVVSLFWPGVGVGWTGSGDDADALLPKGFEGHRLEYTLSQLVPMAAVLGMASCL YLLGVFGKRPTAARRDMSSPQDGVPGITNEKESIGA
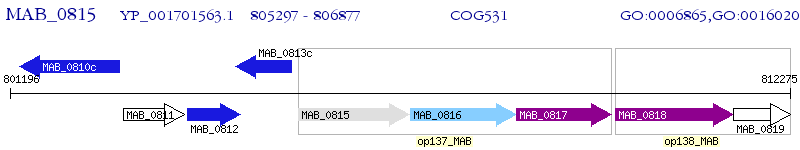
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0815 | - | - | 100% (526) | putative amino acid permease |
| M. abscessus ATCC 19977 | MAB_3599c | - | 2e-14 | 23.66% (465) | putative amino acid permease |
| M. abscessus ATCC 19977 | MAB_3829c | - | 1e-13 | 24.10% (415) | ethanolamine permease |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3281c | - | 2e-14 | 24.53% (424) | cationic amino acid transport integral membrane protein |
| M. gilvum PYR-GCK | Mflv_4126 | - | 2e-14 | 21.35% (431) | amino acid permease-associated region |
| M. tuberculosis H37Rv | Rv3253c | - | 2e-14 | 24.53% (424) | cationic amino acid transport integral membrane protein |
| M. leprae Br4923 | MLBr_01305 | ansP2 | 4e-07 | 22.45% (441) | putative L-asparagine permease |
| M. marinum M | MMAR_1290 | - | 2e-14 | 23.95% (430) | cationic amino acid transport integral membrane protein |
| M. avium 104 | MAV_4216 | - | 2e-16 | 23.11% (463) | cationic amino acid transporter |
| M. smegmatis MC2 155 | MSMEG_2525 | - | 7e-16 | 20.91% (440) | amino acid permease superfamily protein |
| M. thermoresistible (build 8) | TH_0471 | - | 2e-15 | 25.93% (432) | POSSIBLE CATIONIC AMINO ACID TRANSPORT INTEGRAL MEMBRANE |
| M. ulcerans Agy99 | MUL_2597 | - | 1e-14 | 23.72% (430) | cationic amino acid transport integral membrane protein |
| M. vanbaalenii PYR-1 | Mvan_0043 | - | 3e-16 | 25.46% (432) | amino acid permease-associated region |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3281c|M.bovis_AF2122/97 --MAGRRRMKSVEQSIADTDEPTTRLRKDLTWWDLVVFGVSVVIGAGIFT
Rv3253c|M.tuberculosis_H37Rv --MAGRRRMKSVEQSIADTDEPTTRLRKDLTWWDLVVFGVSVVIGAGIFT
MMAR_1290|M.marinum_M --MAGRLRTKSVEQSIADTDEPETRLRKDLTWRDLVVFGVSVVIGAGIFT
MUL_2597|M.ulcerans_Agy99 --MAGGLRTKSVEQSIADTDEPETRLRKDLTWRDLVVFGVSVVIGAGIFT
MAV_4216|M.avium_104 --MASRWRTKSVEQSIADTDEPDTRLRKDLTWWDLVVFGVAVVIGAGIFT
TH_0471|M.thermoresistible__bu --MTDRLRVKSVEQSIADTDEPDSRLRKDLSWWDLTVFGVSVVIGAGIFT
Mflv_4126|M.gilvum_PYR-GCK MTQTEPSPSAGRDKLMTTELVPEQILPKVMTTFGLTAA--YVFIICWITG
MSMEG_2525|M.smegmatis_MC2_155 ---------------MTTELVPEQILPKVMTTFGLTAA--YVFIICWITG
Mvan_0043|M.vanbaalenii_PYR-1 -----------MSISVSPAEQPPQ-LRREFSLGSAFAFAFAFISPIVALY
MAB_0815|M.abscessus_ATCC_1997 --------------MTTSTNTAPSGSGRALNPTLRRVDVFAVLVCALVGV
MLBr_01305|M.leprae_Br4923 MATLAESPEPKSGASRAGVLGEEAGYHKGLKPRQLQMIGIGGAIGTGLFL
: : :.
Mb3281c|M.bovis_AF2122/97 VTASTAGDITGPAIWISFLIAAATCALAALCYAEFASTLPVAGSAYTFSY
Rv3253c|M.tuberculosis_H37Rv VTASTAGDITGPAIWISFLIAAATCALAALCYAEFASTLPVAGSAYTFSY
MMAR_1290|M.marinum_M VTASTAGDITGPAIWISFLIAAGTCALAALCYAEFASTLPVAGSAYTFSY
MUL_2597|M.ulcerans_Agy99 VTASTAGDITGPAIWISFLIAAGTCALAALCYAEFASTLPVAGSAYTFSY
MAV_4216|M.avium_104 VTASTTGDITGPAIWVSFVIAAGTCALAALCYAEFASTLPVAGSAYTFSY
TH_0471|M.thermoresistible__bu VTASTAGNITGPAISVSFIIAAIACGLAALCYAEFASTVPVAGSAYTFSY
Mflv_4126|M.gilvum_PYR-GCK SSVMAGGGWTAIPMWVLGILTFLVP--AGMAVVELGNLWPGQGGVYIWAT
MSMEG_2525|M.smegmatis_MC2_155 SSVMAAGGWTAIPMWVLGILTFLVP--AGMAVVELGNLWPGQGGVYIWAT
Mvan_0043|M.vanbaalenii_PYR-1 GIFGLALSAAGPSFWWGFALVFAGQFLVALVFAALVSRWPLEGSIYQWSR
MAB_0815|M.abscessus_ATCC_1997 DTLGAVSAYGPQGLLWMALVAVIFFLPYGLLTAELGSAFPQEGGPYVWTK
MLBr_01305|M.leprae_Br4923 G-AGGRLAKAGPGLFLVYAVCGVFVFLILRALGELVLHRPSSGSFVSYAR
: : : * *. ::
Mb3281c|M.bovis_AF2122/97 ATFGEFLAWVIGWNLVLELAMGAAVVAKGWSSYLGTVFGFGNGTGHLGSL
Rv3253c|M.tuberculosis_H37Rv ATFGEFLAWVIGWNLVLELAMGAAVVAKGWSSYLGTVFGFGNGTGHLGSL
MMAR_1290|M.marinum_M ATFGEFLAWVIGWNLVLELAIGAAVVAKGWSSYLGTVFGFSGGTADFGSI
MUL_2597|M.ulcerans_Agy99 ATFGEFLAWVIGWNLVLELAIGAAVVAKGWSSYLGTVFGFSGGTADFGSI
MAV_4216|M.avium_104 ATFGEFLAWIIGWNLLLELAIGAAVVAKGWSSYLGTVFGFSGGTVKFGAA
TH_0471|M.thermoresistible__bu ATFGEFVAWIIGWDLILEFSVAAAVVAKGWSSYLGEVFEFAGATTRLGPI
Mflv_4126|M.gilvum_PYR-GCK RTMGETWGFIGGYLSWVPVILNAASSPAVVLQLLLLAFHAELGLTTS---
MSMEG_2525|M.smegmatis_MC2_155 RTMGETWGFIGGYLSWVPVILNAASSPAVVLQLLLLAFHAEIGLTTS---
Mvan_0043|M.vanbaalenii_PYR-1 RLLGSTYGWFAGWVYMWTLVIAMATVALGAAGFVAHIVGIEEASGGT---
MAB_0815|M.abscessus_ATCC_1997 LAFGKFAAGINQFFYWIS-----NPVWIGGTLCIVAVVTFEEFFFPLPGL
MLBr_01305|M.leprae_Br4923 EFFGEKAAYVVGWLYFLDWAMTAIVDTTAIATYLHRWTIFTALPQWT---
:*. . . : :
Mb3281c|M.bovis_AF2122/97 QLDWGALVIVTLVATLIALGTKLSSRFSAVVTAIKVSVVVLVVVVGAFYI
Rv3253c|M.tuberculosis_H37Rv QLDWGALVIVTLVATLIALGTKLSSRFSAVVTAIKVSVVVLVVVVGAFYI
MMAR_1290|M.marinum_M RLDWGALLIVSIVSLLVAYGTKLSSRFSAVVTTIKVAVVILVVVVGAFYV
MUL_2597|M.ulcerans_Agy99 RLDWGALLSVSIVSLLVAYGTKLSSRFSAVVTTIKVAVVILVVVVGAFYV
MAV_4216|M.avium_104 QLDWGALVIVGGVATLIALGTKLSSRFSAVITGIKVSVVVFVVVVGVFYI
TH_0471|M.thermoresistible__bu TLDWGALLIIGFVTVLLATGTKLSANVSLAITVIKVGVVLLVVIVGAFFI
Mflv_4126|M.gilvum_PYR-GCK -IILQLVILWTVIGLALAKLAANQKIMNVVFVVYGVLTLTIFISGLMFAV
MSMEG_2525|M.smegmatis_MC2_155 -IILQLVILWTVIGLALAKLAANQRIMNIVFIVYGVLTLTIFISGLLFAV
Mvan_0043|M.vanbaalenii_PYR-1 -LAWIALLILLAGTAVNLVGRQALKIFMIGSIIAEVVGSVVLGTWLLLFH
MAB_0815|M.abscessus_ATCC_1997 WKWVAAAVFVWGSACVIGASVRFGRWIFLVGAAARIALLGLFIASVIVYA
MLBr_01305|M.leprae_Br4923 ----LALLALAVVLVMNLISVEWFGELEFWAALIKVCALMAFLVVGTIFL
: . : . .
Mb3281c|M.bovis_AF2122/97 RAANYSPFIPEPEVQHHG-GGLDQSVFSLLTGAQGSHYGWYG--VLAGAS
Rv3253c|M.tuberculosis_H37Rv RAANYSPFIPEPEVQHHG-GGLDQSVFSLLTGAQGSHYGWYG--VLAGAS
MMAR_1290|M.marinum_M KAANYSPFIPEPEAGPEG-RGADQSLLSLLTGAPGSHYGWYG--VLAGAS
MUL_2597|M.ulcerans_Agy99 KAANYSPFIPEPEAGPEG-RGADQSLLSLLTGAPGSHYGWYG--VLAGAS
MAV_4216|M.avium_104 KRANYSPFIPKPEAGGQA-KGIDQSVLSLLTGAHTSHYGWYG--VLAGAS
TH_0471|M.thermoresistible__bu KAANYTPFIPPAEAGGEAGRSLDQSLFSLITGAEGSSYGWYG--LLAGAS
Mflv_4126|M.gilvum_PYR-GCK ENGSATPFS--------------------WAEATIPNFAVAG--FLYGT-
MSMEG_2525|M.smegmatis_MC2_155 ENGSATPFS--------------------WAEATIPNFAVAG--FLYGT-
Mvan_0043|M.vanbaalenii_PYR-1 RQNSLSVLFEG-------------------GGTDVDLWSYLGGPFLLAVA
MAB_0815|M.abscessus_ATCC_1997 YKNGLQPMH-----------------------TSDFSWTYAG--FVAVIP
MLBr_01305|M.leprae_Br4923 GGRYPVDGHNTG---------------LSLWTSHGGLFPTGVAPLIVVSS
: : .:
Mb3281c|M.bovis_AF2122/97 IVFFAFIGFDIVATMAEETKRPQRDVPRGILASLGVVTLLYVAVSVVLSG
Rv3253c|M.tuberculosis_H37Rv IVFFAFIGFDIVATMAEETKRPQRDVPRGILASLGVVTLLYVAVSVVLSG
MMAR_1290|M.marinum_M IVFFAFIGFDIVATMAEETKHPQRDVPRGILVSLGVVTLLYVAVSIVLSG
MUL_2597|M.ulcerans_Agy99 IVFFAFIGFDIVATMAEETKHPQRDVPRGILVSLGVVTLLYVAVSIVLSG
MAV_4216|M.avium_104 IVFFAFIGFDIVATMAEETKHPQRDVPRGILASLGIVTLLYVAVAVVLSG
TH_0471|M.thermoresistible__bu IVFFAFIGFDIIATTAEETKNPQRDVARGIMATLGIVTVLYVAVSLVLTG
Mflv_4126|M.gilvum_PYR-GCK -VLLYLLGVETPYNMGAEFLSVRKSGPKMILWGSAALVAIYLLTTLGTMM
MSMEG_2525|M.smegmatis_MC2_155 -VLLYLLGVETPYNMGAEFLSVRKSGPKMILWGSTALVAIYLLTTLGTMM
Mvan_0043|M.vanbaalenii_PYR-1 FIGWSFVGFESAGSIAEEVHNPGRDLPKAVLFSLAFIALVVGYSSLAIIL
MAB_0815|M.abscessus_ATCC_1997 VIVFNFLGFEVPSNAAEEMVNPQKDVPLTVLRGGLTATLLYGIPVLGILL
MLBr_01305|M.leprae_Br4923 GVMFAYAAVELVGTAAGETVEPKKIMPRAINSVIARIAIFYVGSVILLAL
: ..: . . * : . : . . :
Mb3281c|M.bovis_AF2122/97 MVPYTQLRTVPGRGPANLATAFQANG-------------VYWASGIISVG
Rv3253c|M.tuberculosis_H37Rv MVPYTQLRTVPGRGPANLATAFQANG-------------VYWASGIISVG
MMAR_1290|M.marinum_M MVSYTQLRTVSGGGHANLATAFKANG-------------VYWASGIISVG
MUL_2597|M.ulcerans_Agy99 MVSYTQLRTVSGGGHANLATAFKANG-------------VYWASGIISVG
MAV_4216|M.avium_104 MVSYTQLKTMPGRGQANLATAFTANG-------------IHWASKIISIG
TH_0471|M.thermoresistible__bu MVHYTELRAAG--ENANLATAFAANG-------------IDWAAKVISIG
Mflv_4126|M.gilvum_PYR-GCK ALPGDQIDPVTG-----VIGMLDVAG-------------FPGLMEIGAVV
MSMEG_2525|M.smegmatis_MC2_155 ALPGDQIDPVTG-----VIGMLDVAG-------------FPGLMEVGAIV
Mvan_0043|M.vanbaalenii_PYR-1 AIP-DLDAVAEGAVSDPVYETLTTAL-------------GAGIAKPVQVL
MAB_0815|M.abscessus_ATCC_1997 VLPKNQLGTVTGFVDACRAVFTVYGGSVAPDGTAELHGLGQAAAQVAAAG
MLBr_01305|M.leprae_Br4923 LLPYSAFKASES----PFVTFFSKVG-------------FYGAGDLMNIV
:
Mb3281c|M.bovis_AF2122/97 ALAGLTTVVMVLMLGQCRVLFAMARDGLVPRQLAKTGSRGTP--------
Rv3253c|M.tuberculosis_H37Rv ALAGLTTVVMVLMLGQCRVLFAMARDGLVPRQLAKTGSRGTP--------
MMAR_1290|M.marinum_M ALAGLTTVVMVLMLGQCRVLFAMARDGLLPRQLAKTGARGTP--------
MUL_2597|M.ulcerans_Agy99 ALAGLTTVVMVLMLGQCRVLFAMARDGLLPRQLAKTGARGTP--------
MAV_4216|M.avium_104 ALAGLTTVVMVLMLGQCRVLFAMARDGLLPRALAKTGSRGTP--------
TH_0471|M.thermoresistible__bu ALAGLTTVVIVLVLGQTRVLFAMSRDGLLPRPLSKTGPRGTP--------
Mflv_4126|M.gilvum_PYR-GCK LAFIVIVALMTYQVAYSRLIFVSGLERHLPRIFTHLNPRTRNPVTAILIQ
MSMEG_2525|M.smegmatis_MC2_155 LAFIVVVALMTYQVAYSRLIFVSGLERHLPRIFTHLNPRTRNPVTAVLIQ
Mvan_0043|M.vanbaalenii_PYR-1 FVIGFLASFLALQTSASRVIWAYARDGALPAAGTLVQLRGRARIPVVAIL
MAB_0815|M.abscessus_ATCC_1997 LIIGLFTSGTAWAMGVSRAQAVAFTDGAGPAWLGKFSGNGSPARVNIMSA
MLBr_01305|M.leprae_Br4923 VLTAALSSLNAGLYATGRVMHSIAINGSGPKFTARMSKNGVPYGG----I
. . * : * .
Mb3281c|M.bovis_AF2122/97 --VRVTVLVAVLVATTASVFPITKLEEMVNVGTLFAFILVSAGVVVLRRT
Rv3253c|M.tuberculosis_H37Rv --VRVTVLVAVLVATTASVFPITKLEEMVNVGTLFAFILVSAGVVVLRRT
MMAR_1290|M.marinum_M --VRITVLVAVVVGATASVFPITKLEEMVNVGTLFAFVLVSAGVIVLRKK
MUL_2597|M.ulcerans_Agy99 --VRITVLVAVVVGATASVFPITKLEEMVNVGTLFAFVLVSAGVIVLRKK
MAV_4216|M.avium_104 --VRITVLVALVVAVTASVFPITKLEEMVNVGTLFAFVLVSAGVMVLRRT
TH_0471|M.thermoresistible__bu --VRITVLVGALVAIAASVFPVGNLEEMVNIGTLFAFALVSAGVIVLRRT
Mflv_4126|M.gilvum_PYR-GCK GVISSLILVGLYSQSSMANVTVYLQGGLSTVWLLSGFFFLIP-VVVARKK
MSMEG_2525|M.smegmatis_MC2_155 GVISSLILVGLYSQSSMANVTVYLQGGLSTVWLLSGFFFLIP-VIVARKK
Mvan_0043|M.vanbaalenii_PYR-1 VTTVIGAGLFLLSIVAGDIYSLMVNFTAGGFYLAFLFPLIGFLVVVLRKV
MAB_0815|M.abscessus_ATCC_1997 VVSTIVMAAAMIFAHGNAEKYFSAAIGLVVSATAISYILTFSSFIRLRHK
MLBr_01305|M.leprae_Br4923 LLAAVICLCGVALNAFNPGQAFEIVLSVAALGIIAGWGTIVLCQLRLHKM
. : : ::
Mb3281c|M.bovis_AF2122/97 RPD---------LQRGFTAPWVPLLP-----IAAVCACLWLMLN------
Rv3253c|M.tuberculosis_H37Rv RPD---------LQRGFTAPWVPLLP-----IAAVCACLWLMLN------
MMAR_1290|M.marinum_M RPD---------LERGFRAPWVPVLP-----IASLCACLWLMLN------
MUL_2597|M.ulcerans_Agy99 RPD---------LERGFRAPWVPVLP-----IASLCACLWLMLN------
MAV_4216|M.avium_104 RPD---------LERGFKAPWVPVLP-----IASIGACVWLMLN------
TH_0471|M.thermoresistible__bu RPD---------LKRGFRVPFVPWLP-----LAAIAACLWLMLN------
Mflv_4126|M.gilvum_PYR-GCK YADRYESEQFWRIPGGMAGVWTTVVVGS---LATAGGIYYSFVTP-----
MSMEG_2525|M.smegmatis_MC2_155 YADRYANEQFWRIPGGMVGVWITVIVGS---LATVGGIYYSFVTP-----
Mvan_0043|M.vanbaalenii_PYR-1 WKP-----ASFSLRGWTLPVAVVAVVWA---VLQFLNIAWPRVA------
MAB_0815|M.abscessus_ATCC_1997 YPHTR---RPFQVFGGRIGGWIVTVLTVGTMLFTVVSLFWPGVGVGWTGS
MLBr_01305|M.leprae_Br4923 AKAG-------IMRRPRFRMPLAPYSGYLTLAFLFAVLVVMAFDKP----
: .
Mb3281c|M.bovis_AF2122/97 ----------LTALTWIRFGIWLVAGTAIYVGYGRRHSAQGLRQARESAT
Rv3253c|M.tuberculosis_H37Rv ----------LTALTWIRFGIWLVAGTAIYVGYGRRHSAQGLRQARESAT
MMAR_1290|M.marinum_M ----------LTALTWIRFGVWLLVGTVIYVGYGRKHSMVGRRQLAEAAM
MUL_2597|M.ulcerans_Agy99 ----------LTALTWIRFGVWLLVGTVIYVGYGRKHSMVGRRQLAEAAM
MAV_4216|M.avium_104 ----------LTALTWVRFGIWLVVGTAIYAGYGYWHSVQGRRDAGEPDR
TH_0471|M.thermoresistible__bu ----------LTGLTWVRFLGWMALGLVLYFFYGRHHSMLGRGMTDISAR
Mflv_4126|M.gilvum_PYR-GCK ----------WIDVPSSTWMTWVGSISLGMFALGLTVYIFGRRSAHKVSQ
MSMEG_2525|M.smegmatis_MC2_155 ----------WIDVPSATWMTWVGCISLGMFGLGLTVYVFGRRSAHKVSQ
Mvan_0043|M.vanbaalenii_PYR-1 -----------FEQRYLDWSVWIGVVVLGMIGAVLFATVKSRIAAPQDQD
MAB_0815|M.abscessus_ATCC_1997 GDDADALLPKGFEGHRLEYTLSQLVPMAAVLGMASCLYLLGVFGKRPTAA
MLBr_01305|M.leprae_Br4923 ----------IGTWTVASLIVIVPALIAGWYSIRKRVMTIARERMGYTGP
.
Mb3281c|M.bovis_AF2122/97 RRC--------------------
Rv3253c|M.tuberculosis_H37Rv RRC--------------------
MMAR_1290|M.marinum_M DK---------------------
MUL_2597|M.ulcerans_Agy99 DK---------------------
MAV_4216|M.avium_104 SDALAADPNSPVP----------
TH_0471|M.thermoresistible__bu RTG--------------------
Mflv_4126|M.gilvum_PYR-GCK EDSLAHLAVFDLTNKEEEPTK--
MSMEG_2525|M.smegmatis_MC2_155 DDALAHLAVFDLTNQDEAQK---
Mvan_0043|M.vanbaalenii_PYR-1 DRALVDD----------------
MAB_0815|M.abscessus_ATCC_1997 RRDMSSPQDGVPGITNEKESIGA
MLBr_01305|M.leprae_Br4923 FPAIANPPVQPSERSHSQNP---