For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSQSTGQFRAHHQSTSQDWAALSAWVARQGHTLETSGARQFAGGKANLNYLVVLDGQPAVFRRPPAGPIA EGANDMAREWRVLSRLGDGFALAPRGLLYCDDLDVLAAPFQFLEYREGLAIGGELPAGLPVDAPSRITAT LIEAMTALHRVSPERVGLGELGRPEGLLERQLTGWTRRAHVVWPDGLPDVVTYLTSRLAEEIPEPSAISL LHMDLKLDNMLIDQETLSPNAIIDWDMATRGCPLFDLAILVSYWMEPDDPAGVRAVNQMPTTAQGFGKRA DVIDSYFAAAGMPPRPLHWHVACARLRLAVAWMQLYRKWQSGDLIGPGYADFENIAMSVLDWATTQFTEG EI
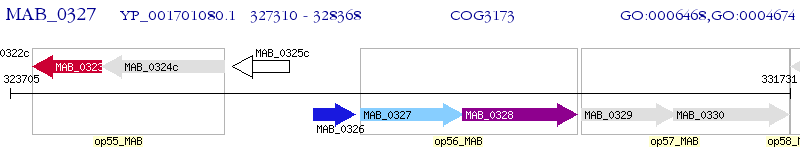
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0327 | - | - | 100% (352) | aminoglycoside phosphotransferase |
| M. abscessus ATCC 19977 | MAB_4589c | - | 9e-32 | 29.68% (310) | putative phosphotransferase |
| M. abscessus ATCC 19977 | MAB_4910c | - | 3e-17 | 25.72% (311) | putative aminoglycoside phosphotransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3787c | fadE36 | 1e-32 | 32.73% (275) | acyl-CoA dehydrogenase |
| M. gilvum PYR-GCK | Mflv_1204 | - | 7e-35 | 34.63% (283) | aminoglycoside phosphotransferase |
| M. tuberculosis H37Rv | Rv3761c | fadE36 | 1e-32 | 32.73% (275) | acyl-CoA dehydrogenase |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_0155 | fadE36_1 | 8e-33 | 32.98% (285) | acyl-CoA dehydrogenase FadE36_1 |
| M. avium 104 | MAV_0316 | - | 4e-33 | 33.22% (301) | phosphotransferase enzyme family protein |
| M. smegmatis MC2 155 | MSMEG_6337 | - | 5e-33 | 37.56% (221) | phosphotransferase enzyme family protein |
| M. thermoresistible (build 8) | TH_1657 | fadE36 | 5e-33 | 34.69% (294) | POSSIBLE ACYL-CoA DEHYDROGENASE FADE36 |
| M. ulcerans Agy99 | MUL_4942 | fadE36_1 | 9e-33 | 32.98% (285) | acyl-CoA dehydrogenase FadE36_1 |
| M. vanbaalenii PYR-1 | Mvan_5603 | - | 2e-31 | 32.53% (292) | aminoglycoside phosphotransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1204|M.gilvum_PYR-GCK ----------MTELEGLDLAALDRHLREVGVPRSGELRAELIAGGRSNLT
Mvan_5603|M.vanbaalenii_PYR-1 ----------MTELEGLDLAALDRHLCDVGVPRSGELRAEFISGGRSNLT
TH_1657|M.thermoresistible__bu --------MAVTSLDGLDLDALDRHLRQVGVPRSGELRAELIAGGRSNLT
MSMEG_6337|M.smegmatis_MC2_155 ----------MTSLEGLDLDALDRHLRAEGIARAGDLRAELIAGGRSNLT
Mb3787c|M.bovis_AF2122/97 -------MTSVDRLDGLDLGALDRYLRSLGIGRDGELRGELISGGRSNLT
Rv3761c|M.tuberculosis_H37Rv -------MTSVDRLDGLDLGALDRYLRSLGIGRDGELRGELISGGRSNLT
MAV_0316|M.avium_104 -------MTSADQLEGLDLAALDSYLRSLGIGRDGELRAEFISGGRSNLT
MMAR_0155|M.marinum_M -----MPGPMSEDIPGVHAEAVSRWLVTLGHQVAAPLRFQRIGLGQSNLT
MUL_4942|M.ulcerans_Agy99 ---------MSEDIPGVHAHAVSRWLVTLGHQVAAPLRFQRIGLGQANLT
MAB_0327|M.abscessus_ATCC_1997 MSQSTGQFRAHHQSTSQDWAALSAWVARQGHTLETSG-ARQFAGGKANLN
. . *:. : * . :. *::**.
Mflv_1204|M.gilvum_PYR-GCK FLVGDDA-SEWVLRRPPLHGLTPSAHDMAREYKVVAALAGTKVPVARAVT
Mvan_5603|M.vanbaalenii_PYR-1 FLVFDDA-SKWVLRRPPLHGLTPSAHDMAREYRVVAALADTAVPVARAVT
TH_1657|M.thermoresistible__bu FLVFDDQ-SRWVLRRPPLHGLTPSAHDMAREYRVVAALADTPVPVARAVT
MSMEG_6337|M.smegmatis_MC2_155 FLVFDDA-SKWVLRRPPLHGLTPSAHDMAREYRVVAALAGTPVPVARAVT
Mb3787c|M.bovis_AF2122/97 FRVYDDA-SSWLVRRPPLHGLTPSAHDMAREYRVVAALGDTPVPVARTIS
Rv3761c|M.tuberculosis_H37Rv FRVYDDA-SSWLVRRPPLHGLTPSAHDMAREYRVVAALGDTPVPVARTIS
MAV_0316|M.avium_104 FRVYDDA-TSWLVRRPPLHGLTPSAHDMAREYRVVAALQDTPVPVARTIG
MMAR_0155|M.marinum_M YRVTDAAGRAWVLRRPPLGHLLASAHDVVREARIMAALAETDVPVPRILG
MUL_4942|M.ulcerans_Agy99 YRVTDAAGRAWVLRHPPLGHLLASAHDVVREARIMAALAETDVPVPRILG
MAB_0327|M.abscessus_ATCC_1997 YLVVLDG-QPAVFRRPPAGPIAEGANDMAREWRVLSRLGDGFALAPRGLL
: * :.*:** : .*:*:.** :::: * . ..* :
Mflv_1204|M.gilvum_PYR-GCK MSDDDSVLGAPFQMVERVAGLVVRHTPQLAALGDETVISNVVDSLIQVLA
Mvan_5603|M.vanbaalenii_PYR-1 MRNDDSVLGAPFQMVEYVAGRVVRHTDQLAALGDRATIEKCVDALIKVLA
TH_1657|M.thermoresistible__bu MSNDDSVLGAPFQMVEHVPGLVVRYTDDLKSVGDRDAIEACVDSLIRVLA
MSMEG_6337|M.smegmatis_MC2_155 MRNDDSVLGAPFQMVDHVDGRVVRTAAELAALGDQTVIDNCIDALIKALS
Mb3787c|M.bovis_AF2122/97 LCQDDSVLGAPFQVVEFVAGQVVRRRAELEALGSRSVIEGCVDALIRVLV
Rv3761c|M.tuberculosis_H37Rv LCQDDSVLGAPFQVVEFVAGQVVRRRAELEALGSRSVIEGCVDALIRVLV
MAV_0316|M.avium_104 LCEDESVLGAPFQIVEFVAGQVVRRRAQLESF-SHTVIEGCVDSLIRVLV
MMAR_0155|M.marinum_M VTTDSEFSEAPLVLMEFVDGLVVDTMEVAEALTPRQRRQ-IALSLPRTLA
MUL_4942|M.ulcerans_Agy99 VTTDSEFSEAPLVLMEFVDGLVVDTMEVAEALTPRQRRQ-IALSLPRTLA
MAB_0327|M.abscessus_ATCC_1997 YCDDLDVLAAPFQFLEYREGLAIG--GELPAGLPVDAPSRITATLIEAMT
* .. **: .:: * .: : . :* ..:
Mflv_1204|M.gilvum_PYR-GCK DLHEVDPESVGLGDFGKASGYLERQVRRWGSQWDLVKRDDDPSDADVRKL
Mvan_5603|M.vanbaalenii_PYR-1 DLHSVDPDAVGLSDFGKPSGYLERQVRRWGSQWDLVRRADDPCDADVRRL
TH_1657|M.thermoresistible__bu DLHAVDYEAVGLGDFGKPSGYLERQVRRWASQWELVKHPDDARDGDVKKL
MSMEG_6337|M.smegmatis_MC2_155 DLHAVDPYAVGLGDFGKPDGYLERQVRRWGSQWEHVRLPDDARDDDVRRL
Mb3787c|M.bovis_AF2122/97 DLHSIDPKAVGLSDFGKPDGYLERQVRRWGSQWELVRLPDDHRDADISRL
Rv3761c|M.tuberculosis_H37Rv DLHSIDPKAVGLSDFGKPDGYLERQVRRWGSQWELVRLPDDHRDADISRL
MAV_0316|M.avium_104 DLHSVDPDAVGLADFGKPSGYLERQVRRWGSQWALVRLPEDRRDADVERL
MMAR_0155|M.marinum_M KIHAVDLAKVGLDDLASHKPYAQRQLKRWAGQWELSKTRELP---ELDDL
MUL_4942|M.ulcerans_Agy99 KIHAVDLAKVGLDDLASHKPYAQRQLKRWAGQWELSKTRELP---ELDDL
MAB_0327|M.abscessus_ATCC_1997 ALHRVSPERVGLGELGRPEGLLERQLTGWTRRAHVVWPDGLP--DVVTYL
:* :. *** ::. . :**: * : : *
Mflv_1204|M.gilvum_PYR-GCK HQRLAESVPEQSRASIVHGDYRIDNTILDADDPTKVAAVLDWEMSTLGDP
Mvan_5603|M.vanbaalenii_PYR-1 HERLSAALPPQSRSSIVHGDYRIDNTILDAEDPTKVVAVLDWEMSTLGDP
TH_1657|M.thermoresistible__bu HARLEQAVPPQSRTSIVHGDYRIDNTILDADDPTEVVAVLDWEMSTLGDP
MSMEG_6337|M.smegmatis_MC2_155 HAALADAVPPQSGTSIVHGDYRIDNTVLDAEDATVVRAVLDWEMSTLGDP
Mb3787c|M.bovis_AF2122/97 HLALQQAIPQQSRTSIVHGDYRIDNTILDTDDPCHVRAVVDWELSTLGDP
Rv3761c|M.tuberculosis_H37Rv HLALQQAIPQQSRTSIVHGDYRIDNTILDTDDPCHVRAVVDWELSTLGDP
MAV_0316|M.avium_104 HSGLGQAIPQQSRTSIVHGDYRIDNTILDADDPTKVRAVVDWELSTLGDP
MMAR_0155|M.marinum_M TRRLVAAAPEQREISLVHGDFHLRN-LITSPETGAVVATLDWELSTLGDP
MUL_4942|M.ulcerans_Agy99 TRRLVAAAPEQREISLVHGDFHLRN-LITSPETGAVVATLDWELSTLGDP
MAB_0327|M.abscessus_ATCC_1997 TSRLAEEIPEPSAISLLHMDLKLDNMLIDQETLSPN-AIIDWDMATRGCP
* * *::* * :: * :: * :**:::* * *
Mflv_1204|M.gilvum_PYR-GCK LSDAALMCVYRLP---AFN-EVHDDAAWACDLMPPTDDLAQRYALASGKP
Mvan_5603|M.vanbaalenii_PYR-1 LSDAALMCVYRHP---MFN-MVHADAAWASELIPDADALADKYSRAAGQP
TH_1657|M.thermoresistible__bu LSDAALMCVYRHP---TFN-YVHSDAAWASELIPSADALAEKYSTAAGQD
MSMEG_6337|M.smegmatis_MC2_155 LSDAALMCVYRNP---MFG-HIHTDAAWSSDLVPPADELADRYSKVSGRD
Mb3787c|M.bovis_AF2122/97 LSDAALMCVYRDP---ALDLIVHAQAAWTSPLLPAADELADRYSLVSGQP
Rv3761c|M.tuberculosis_H37Rv LSDAALMCVYRDP---ALDLIVHAQAAWTSPLLPAADELADRYSLVSGQP
MAV_0316|M.avium_104 LSDAALMCVYRDP---ALDLIVNAQAAWTSPLLPTADELADRYSLVAGIP
MMAR_0155|M.marinum_M LADMGSLLTYWME---PGESGVGDFAPSTLDGFPDRAEMTRLYLEETGRD
MUL_4942|M.ulcerans_Agy99 LADMGSLLTYWME---PGESGVGDFAPSTLDGFPDRAEMTRLYLEETGRD
MAB_0327|M.abscessus_ATCC_1997 LFDLAILVSYWMEPDDPAGVRAVNQMPTTAQGFGKRADVIDSYFAAAGMP
* * . : * . : . : * :*
Mflv_1204|M.gilvum_PYR-GCK LDHWDFYMALGYFKIAIIAAGIAYRARMGGGV---AEGTDEVDRAVRPFV
Mvan_5603|M.vanbaalenii_PYR-1 LDHWDFYMALAYFKLAIIGAGIAYRAREAG-V---VDDTDKVGEAVAPLI
TH_1657|M.thermoresistible__bu LVHWDFYMALAYFKLAIIAAGIDYRARESG-A---GGSTDRVGEAVAPLI
MSMEG_6337|M.smegmatis_MC2_155 LAHWDFYMALAYFKLAIIAAGIDYRRREGA------DGPEQVGEAVAPMI
Mb3787c|M.bovis_AF2122/97 LGHWEFYMALAYFKLAIIAAGIDYRRRMSEQAEGKDTAAESVPDVVAPLI
Rv3761c|M.tuberculosis_H37Rv LGHWEFYMALAYFKLAIIAAGIDYRRRMSEQAEGKDTAAESVPDVVAPLI
MAV_0316|M.avium_104 LAHWEFYMALAYFKLAIIAAGIDFRRRMSDQARGLGEAAEHTPEVVAPLI
MMAR_0155|M.marinum_M PAALQYWHVLGLWKLAIIAEGVMRRAMDEPQNKASEGTPTVAWIDARVHM
MUL_4942|M.ulcerans_Agy99 PAALQYWHVLGLWKLAIIAEGVMRRAMDEPQNKASEGTPTVAWIDARVHM
MAB_0327|M.abscessus_ATCC_1997 PRPLHWHVACARLRLAVAWMQLYRKWQSGDLIGPGYADFENIAMSVLDWA
.: . . ::*: : : .
Mflv_1204|M.gilvum_PYR-GCK AAGLHILS----------
Mvan_5603|M.vanbaalenii_PYR-1 AAGLSQLA----------
TH_1657|M.thermoresistible__bu DAGLRTLRA---------
MSMEG_6337|M.smegmatis_MC2_155 AAGLAALG----------
Mb3787c|M.bovis_AF2122/97 ARGLAEIAKKSG------
Rv3761c|M.tuberculosis_H37Rv ARGLAEIAKKSG------
MAV_0316|M.avium_104 SRGLAELAKLPG------
MMAR_0155|M.marinum_M AREIADAAGMNQVSDSAS
MUL_4942|M.ulcerans_Agy99 AREIADAAGMNQVSDSAF
MAB_0327|M.abscessus_ATCC_1997 TTQFTEGEI---------
: