For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MRTRDQIKYWSRWATMHGISRAALLTQVRTLPLAALFLGPDRAEKHYRYIEEIRASGSVTPSRAGGLIFT DLALTREILRDNRFITMAPTNIPSPILPLAFSRWVFARTEPGLPNPVEPPAMLAVDPPEHTRFRKLVSKA FTPRAVSKLEDRVREVTTELLDNLERHERADLLHDYASQLPVAIIAEMLGVPRADAPFLLEWGNHGAALL DIGMTWSAYRDATQALIEIDRYFDAHLVRLRGELAEDPTVDGILASIVRDGDLNDRELKATMALLLGAGF ETTVNLIGNGIAALLRHPQQLAHLRENPDGWPNAVEEILRYDSPVQITGRVATQACEFEGHTLAAGSMAI LLLGGANRDPAVFDQPDVFDVLRANAREHVAFGSGIHVCLGASLARMEGVVALQSLFERFPELALAADPT PGKHVNLHGFGSLPVNLGRARVPASS
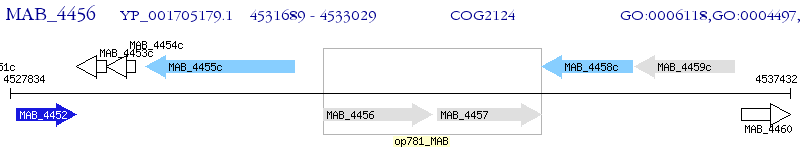
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4456 | - | - | 100% (446) | putative cytochrome P450 |
| M. abscessus ATCC 19977 | MAB_4457 | - | e-102 | 47.02% (436) | putative cytochrome P450 |
| M. abscessus ATCC 19977 | MAB_0276 | - | 7e-53 | 38.44% (333) | cytochrome P450 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1912c | cyp140 | 9e-86 | 42.20% (436) | cytochrome p450 140 CYP140 |
| M. gilvum PYR-GCK | Mflv_0288 | - | 8e-93 | 43.22% (435) | cytochrome P450 |
| M. tuberculosis H37Rv | Rv1880c | cyp140 | 9e-86 | 42.20% (436) | cytochrome p450 140 CYP140 |
| M. leprae Br4923 | MLBr_02088 | - | 4e-56 | 39.94% (333) | putative cytochrome p450 |
| M. marinum M | MMAR_2768 | cyp140A5 | 1e-87 | 43.31% (441) | cytochrome P450 140A5 Cyp140A5 |
| M. avium 104 | MAV_2156 | - | 1e-115 | 49.10% (446) | P450 heme-thiolate protein |
| M. smegmatis MC2 155 | MSMEG_0681 | - | 2e-92 | 43.91% (419) | P450 heme-thiolate protein |
| M. thermoresistible (build 8) | TH_2999 | - | 2e-87 | 41.00% (439) | PUTATIVE cytochrome P450 |
| M. ulcerans Agy99 | MUL_2979 | cyp140A5 | 5e-88 | 43.54% (441) | cytochrome P450 140A5 Cyp140A5 |
| M. vanbaalenii PYR-1 | Mvan_5525 | - | 1e-118 | 51.92% (443) | cytochrome P450 |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_2156|M.avium_104 MRVR----LGARWMAMHGLPRAYFAVQARR-----GDPLARLLRSGTTGE
Mvan_5525|M.vanbaalenii_PYR-1 MRSRP--AVWLRWATVHGVPRAFLTVRARR-----GEPLARLILG---RG
MAB_4456|M.abscessus_ATCC_1997 MRTRDQIKYWSRWATMHGISRAALLTQVR------TLPLAALFLGPDRAE
Mflv_0288|M.gilvum_PYR-GCK -MAVHPLRQRIRWLALHGVIRGLSKLAARRS----GDPQARLIADPEARN
MSMEG_0681|M.smegmatis_MC2_155 ----------------------MATLGLRR-----GDPQARLIADPVVRA
TH_2999|M.thermoresistible__bu -------------LSLHGVIRALTAYGARR-----GDPQARLIADPAVRA
Mb1912c|M.bovis_AF2122/97 ------MKDKLHWLAMHGVIRGIAAIGIRR-----GDLQARLIADPAVAT
Rv1880c|M.tuberculosis_H37Rv ------MKDKLHWLAMHGVIRGIAAIGIRR-----GDLQARLIADPAVAT
MMAR_2768|M.marinum_M ------MKDKLHWFAMHGVIRGMANIGLRR-----GELEARLIADPAVAA
MUL_2979|M.ulcerans_Agy99 ------MKDKLHWFAMHGVIRGMANIGLRR-----GELEARLIADPAVAA
MLBr_02088|M.leprae_Br4923 --MRTCVPTRTCVYAFIEYLSHNRPMGTNPPSLVEAQMLLLRLIDPGTRA
. : .
MAV_2156|M.avium_104 DRYALMEQIRARGPLMRAPF-VWASVDHAVCRQVLRDKRFGVTSPTEMEL
Mvan_5525|M.vanbaalenii_PYR-1 DRLALIEEIRAAGPLMRTPV-VWATADYDVCRTVLRDNDFGVADPSETGL
MAB_4456|M.abscessus_ATCC_1997 KHYRYIEEIRASGSVTPSRAGGLIFTDLALTREILRDNRFITMAPTNIPS
Mflv_0288|M.gilvum_PYR-GCK DPAAFADEVRTGDPVVRCRA-VLLTVDHQAANEILRSDDFRVSSLG-AGL
MSMEG_0681|M.smegmatis_MC2_155 DPTTFADQMRKQGPVIRGRV-VLLTFDHAVAFDLLRSDDFQVSQLG-ANL
TH_2999|M.thermoresistible__bu DPAAFADELRGRGPLIRGRA-ALLTFDHGVANELLRSDDFRVSELG-GNL
Mb1912c|M.bovis_AF2122/97 DPVPFYDEVRSHGALVRNRA-NYLTVDHRLAHDLLRSDDFRVVSFG-ENL
Rv1880c|M.tuberculosis_H37Rv DPVPFYDEVRSHGALVRNRA-NYLTVDHRLAHDLLRSDDFRVVSFG-ENL
MMAR_2768|M.marinum_M NPGPFHDELRRHGRMVRTRV-NYLTVDYQLAHDVLRSDDFHTISFG-ENL
MUL_2979|M.ulcerans_Agy99 NPGPFHDELRRHGRMVRTRV-NYLTVDYQLAHDVLRSDDFHTISFG-ENL
MLBr_02088|M.leprae_Br4923 DPFPVYRALIDYGPMQLPGMPLTVFSSFSDCDEALRHPLSASDRLKATLA
. : . : . **
MAV_2156|M.avium_104 PRPVRAL----IARTDPGVANPVAPPAMVIVDPPDHTRYRQLVAQSFTPR
Mvan_5525|M.vanbaalenii_PYR-1 PDALLGL----IRRVDPGLPNPVEPPAMLMTDPPQHTEYRKLVSRSFTPR
MAB_4456|M.abscessus_ATCC_1997 PILPLAFSRWVFARTEPGLPNPVEPPAMLAVDPPEHTRFRKLVSKAFTPR
Mflv_0288|M.gilvum_PYR-GCK PAPLRWV----NRKTHPGLLHPIEPPSLLSIEPPDHTRCRKLVSSVFTTR
MSMEG_0681|M.smegmatis_MC2_155 PKPLRWI----ADKTNPGLLHPIEPPSLLSIEPPDHTRMRKLVSSVFTTR
TH_2999|M.thermoresistible__bu PAPLRWV----AAKADTGHLHPIQPPSLLSIEPPDHTRLRKLVSSVFTTR
Mb1912c|M.bovis_AF2122/97 PPPLRWL----ERRTRGDQLHPLREPSLLAVEPPDHTRYRKTVSAVFTSR
Rv1880c|M.tuberculosis_H37Rv PPPLRWL----ERRTRGDQLHPLREPSLLAVEPPDHTRYRKTVSAVFTSR
MMAR_2768|M.marinum_M PGPLRWL----ERRTRDEQLHPLRPPSLLAVEPPDHTRYRKTVSAVFTSR
MUL_2979|M.ulcerans_Agy99 PGPLRWL----ERRTRDEQLHPLRPPSLLAVEPPDHTRYRKTVSAVFTSR
MLBr_02088|M.leprae_Br4923 QQAIAAG----------AEPRPFYASSFMFLDPPDHTRLRKLVSKAFAPK
.*. .::: :**:**. *: *: *:.:
MAV_2156|M.avium_104 AIEALNTRVAQVTMELIERIAAT---PQPDLIADFATRLPVAIIAEILGM
Mvan_5525|M.vanbaalenii_PYR-1 SIAALDTRIGELTAELLDDLGRR---REVDLIADYAAQLPAAVISRMLGV
MAB_4456|M.abscessus_ATCC_1997 AVSKLEDRVREVTTELLDNLERH---ERADLLHDYASQLPVAIIAEMLGV
Mflv_0288|M.gilvum_PYR-GCK AVNALRDRVQLTADQLLDDLERE--SGVVDIVERYCSQLPVAVISDILGV
MSMEG_0681|M.smegmatis_MC2_155 AVATLRERVEQTAHELLDGLEAE--SGVVDIVERYCAQLPVAVIGDILGV
TH_2999|M.thermoresistible__bu AVATLKERLQQTAFDLLDGLDEVGASDVVDVIDRYCSQLPIAVISDILGV
Mb1912c|M.bovis_AF2122/97 AVSALRDLVEQTAINLLDRFAEQ--PGIVDVVGRYCSQLPIVVISEILGV
Rv1880c|M.tuberculosis_H37Rv AVSALRDLVEQTAINLLDRFAEQ--PGIVDVVGRYCSQLPIVVISEILGV
MMAR_2768|M.marinum_M AVAALRDRVEQTANSLLDQLAQR--PGVVDVVGQYCSQLPIMVISEILGV
MUL_2979|M.ulcerans_Agy99 AVAALRDRVEQTANSLLDQLAQR--PGVVDVVGQYCSQLPIMVISEILGV
MLBr_02088|M.leprae_Br4923 VVQALEGDIAALVDSLLDKGAAAG---QFDVIADLAFPLAVAVICRLLGV
: * : . .*:: *:: . *. :* :**:
MAV_2156|M.avium_104 PPDSYPRMLAWGRSGSPLLDLGIDWK-TYRDAIAGLRGVDEYLLAHFHQL
Mvan_5525|M.vanbaalenii_PYR-1 PDEDRARILGWGETGAALLDIGIGWR-PFRAAIDGLVDVDTHLGAHFRRL
MAB_4456|M.abscessus_ATCC_1997 PRADAPFLLEWGNHGAALLDIGMTWS-AYRDATQALIEIDRYFDAHLVRL
Mflv_0288|M.gilvum_PYR-GCK PEQDRKHILHFGELGAPSLDIGLSWR-QFQQVEEGLVGFNAWLGDHLEAL
MSMEG_0681|M.smegmatis_MC2_155 PEPDRPNILRYGELGAPSLDIGLSWS-QYQQVNEGIIGFETWLNRHLRTL
TH_2999|M.thermoresistible__bu PDEDRDRILHFGELGAPSLDIGLSWR-QYRQVREGIVGFNTWLTAHLQRL
Mb1912c|M.bovis_AF2122/97 PEHDRPRVLEFGELAAPSLDIGIPWR-QYLRVQQGIRGFDCWLEGHLQQL
Rv1880c|M.tuberculosis_H37Rv PEHDRPRVLEFGELAAPSLDIGIPWR-QYLRVQQGIRGFDCWLEGHLQQL
MMAR_2768|M.marinum_M PEQDRGRVLEYGELAAPSLDIGVPYR-QYRRIQQGITGFNSWLTAHLEQL
MUL_2979|M.ulcerans_Agy99 PEQDRGRVLEYGELAAPSLDIGVTYR-QYRRIQQGITGFNSWLTAHLEQL
MLBr_02088|M.leprae_Br4923 PYEDAPEFGRVSALLVQSVDPFITITGEPPEATEERLRAGVWLRDYLEQL
* . . . :* : : :: *
MAV_2156|M.avium_104 R----ADPHSDNPFGRMAADGS---LTDRELTANAALIVGAGFETTVNLI
Mvan_5525|M.vanbaalenii_PYR-1 H----QNGADGTPFSSLALDGS---LSGRELISNAALLVGAGVETTVNLI
MAB_4456|M.abscessus_ATCC_1997 RGELAEDPTVDGILASIVRDGD---LNDRELKATMALLLGAGFETTVNLI
Mflv_0288|M.gilvum_PYR-GCK RRNPGDDLMSELILASAAADEAG-RLSERELQATAGLVLAAGFETTVNLL
MSMEG_0681|M.smegmatis_MC2_155 RRDPGDDLLSQIITASESGATGE-PLTDRELKALAGLVLAAGFETTVNLL
TH_2999|M.thermoresistible__bu RRDPGEDLLSQLIQAADAETT------ERELEALAGLVLAAGFETTVNLL
Mb1912c|M.bovis_AF2122/97 RHAPGDDLMSQLIQIAESGDNET-QLDETELRAIAGLVLVAGFETTVNLL
Rv1880c|M.tuberculosis_H37Rv RHAPGDDLMSQLIQIAESGDNET-QLDETELRAIAGLVLVAGFETTVNLL
MMAR_2768|M.marinum_M RRNPGDDLMSQLIRVAESGDAGT-YLDESELRAVAGLVLVAGFETTVNLL
MUL_2979|M.ulcerans_Agy99 RRNPGDDLMSQLIRVAESGDAGT-YLDKSELRAIAGLVLVAGFETTVNLL
MLBr_02088|M.leprae_Br4923 VKCRRGTPGEDLISRLIELDESGDQLTEEEIIATCGLLLVAGHETTVNLI
*: : .*:: ** ******:
MAV_2156|M.avium_104 GNGIVLLLRHPEQLALLHDNPDLWPSAVEEILRIASPVQMTARTPACDVD
Mvan_5525|M.vanbaalenii_PYR-1 GNGIAALLDHPDQLALLRDDPGLWPSAVEEILRYDSPVQMTARTAHRDTE
MAB_4456|M.abscessus_ATCC_1997 GNGIAALLRHPQQLAHLRENPDGWPNAVEEILRYDSPVQITGRVATQACE
Mflv_0288|M.gilvum_PYR-GCK GNGIRMLLDTPQHLETLAARPDLWPNTVEEILRLDSPVQMSARFARRDVE
MSMEG_0681|M.smegmatis_MC2_155 GNGIRMLLDHREHLETLAARPDVWPNAVEEILRLDSPVQMTARAAVRDVD
TH_2999|M.thermoresistible__bu GNGIRMLLDTPGHLDTLAARPELWPNAVEEILRLDPPVQMTARVARVDTD
Mb1912c|M.bovis_AF2122/97 GNGIRMLLDTPEHLATLRQHPELWPNTVEEILRLDSPVQLTARVACRDVE
Rv1880c|M.tuberculosis_H37Rv GNGIRMLLDTPEHLATLRQHPELWPNTVEEILRLDSPVQLTARVACRDVE
MMAR_2768|M.marinum_M GNGIRMLLDAPEQLETLRQRPELWPNAVEEILRLDSPVQLTARVARTDVE
MUL_2979|M.ulcerans_Agy99 GNGIRMLLDAPEQLETLRQRPELWPNAVEEILRLDSPVQLTARVARTDVE
MLBr_02088|M.leprae_Br4923 ANAVLAMLRNPSQWKALSSNPQRAPLVVEETLRYDPAIHLIGRVAAKDMT
.*.: :* : * * * .*** ** ..::: .* .
MAV_2156|M.avium_104 IAGAHIGAGEMVGLFLGGANRDPKVFSDPTTFDVTRPNAREHLAFASGIH
Mvan_5525|M.vanbaalenii_PYR-1 IAGVGIRKGAVIVMLLGGANRDPKVFDRPQTFDITRANARDHLAFASGIH
MAB_4456|M.abscessus_ATCC_1997 FEGHTLAAGSMAILLLGGANRDPAVFDQPDVFDVLRANAREHVAFGSGIH
Mflv_0288|M.gilvum_PYR-GCK VAGTRIERGELVIVHLAGANRDPNVFDDPHRFDIERDNAGRHLSFSGGRH
MSMEG_0681|M.smegmatis_MC2_155 VAGTAVGRGELVVLHIAGANRDPKVFDDPHRFDIERDNAGRHLSFSGGRH
TH_2999|M.thermoresistible__bu IAGTAVRRGEMVIIHLAGANRDPKVFADPHRFDIERANAHRHLSFSGGRH
Mb1912c|M.bovis_AF2122/97 VAGVRIKRGEVVVIYLAAANRDPAVFPDPHRFDIERPNAGRHLAFSTGRH
Rv1880c|M.tuberculosis_H37Rv VAGVRIKRGEVVVIYLAAANRDPAVFPDPHRFDIERPNAGRHLAFSTGRH
MMAR_2768|M.marinum_M IASWPINRGEMVVAYLAGANRDPAVFPNPHRFDVERANAGRHLAFSTGRH
MUL_2979|M.ulcerans_Agy99 IASWPINRGEMVVAYLAGANRDPAVFPNPHRFDVERANAGRHLAFSTGRH
MLBr_02088|M.leprae_Br4923 IGQTTLTEGDTMVLLLAAANRDPAVYSRPDEFDPDRPSS-RHLAFAVGSH
. : * :..***** *: * ** * .: *::*. * *
MAV_2156|M.avium_104 ACLGAALARIEGATALRALFENFPDLRLTAAPQRRSLINLHGYTRLPAQL
Mvan_5525|M.vanbaalenii_PYR-1 VCLGAALARIEGATALRELFTRYPDLHLTAPAEPRDLVTLHGYRHLPVRL
MAB_4456|M.abscessus_ATCC_1997 VCLGASLARMEGVVALQSLFERFPELALAADPTPGKHVNLHGFGSLPVNL
Mflv_0288|M.gilvum_PYR-GCK FCLGAALARAEGEVGLRTFFQRYPDAQLAGSGSRRDTRVLRGWSTLPITL
MSMEG_0681|M.smegmatis_MC2_155 FCLGAALARAEGEVGLRAFFDRFPDARLAGDPVRRDTRVLRGWSTLPIAL
TH_2999|M.thermoresistible__bu FCLGAALARAEGEVGLRTFFQRYPKARSAGGGDRRDTRVLRGWASLPITL
Mb1912c|M.bovis_AF2122/97 FCLGAALARAEGEVGLRTFFDRFPDVRAAGAGSRRDTRVLRGWSTLPVTL
Rv1880c|M.tuberculosis_H37Rv FCLGAALARAEGEVGLRTFFDRFPDVRAAGAGSRRDTRVLRGWSTLPVTL
MMAR_2768|M.marinum_M FCLGAALARAEGEVGLRTFFERFPDVRAAGAGSRRDTRVLRGWSTLPVTL
MUL_2979|M.ulcerans_Agy99 FCLGAALARAEGEVGLRTFFERFPDVRAAGAGSRRDTRVLRGWSTLPVTL
MLBr_02088|M.leprae_Br4923 FCLGAALARLEATVTLSAISARFPQVQLAGELVYKPNVAMRGMSALPVQV
****:*** *. . * : .:*. :. ::* ** :
MAV_2156|M.avium_104 GGRRTTSTTIPV
Mvan_5525|M.vanbaalenii_PYR-1 G-RAATATRRAS
MAB_4456|M.abscessus_ATCC_1997 GRARVPASS---
Mflv_0288|M.gilvum_PYR-GCK GKAPAALGS---
MSMEG_0681|M.smegmatis_MC2_155 GKARAAVGS---
TH_2999|M.thermoresistible__bu GPARTTVGS---
Mb1912c|M.bovis_AF2122/97 GPARSMVSP---
Rv1880c|M.tuberculosis_H37Rv GPARSMVSP---
MMAR_2768|M.marinum_M GPARALAAS---
MUL_2979|M.ulcerans_Agy99 GPARALAAS---
MLBr_02088|M.leprae_Br4923 ------------