For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTGSRGAGQQRARVLKVLQEAAAPVGAREVADSLGLHITTVRFHLKTLEEQGHIVRRGGTGEQRAGRPSL SYAVAPRLDYADVVALFAVHLGGTAAERESRAALVGADLAHRVNVARRRTPLQVTDLVVETLGELGFTVR STLMSFGRVTVQICSCPLAEIATTAPEVVRGIQRGLIQEVLDVNADAIGGQFQVTVSPDAGHGDCTVNLA LVPLGKESWPWT
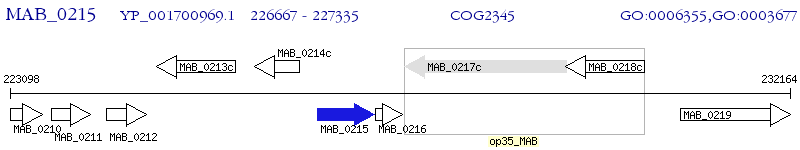
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0215 | - | - | 100% (222) | transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_2750c | - | 2e-05 | 29.38% (194) | transcriptional regulatory protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2654c | - | 2e-14 | 32.32% (198) | transcriptional regulatory protein |
| M. gilvum PYR-GCK | Mflv_4298 | - | 3e-71 | 65.07% (209) | putative transcriptional regulator |
| M. tuberculosis H37Rv | Rv2621c | - | 2e-13 | 34.81% (181) | transcriptional regulatory protein |
| M. leprae Br4923 | MLBr_00592 | - | 4e-06 | 24.77% (218) | putative DNA-binding protein |
| M. marinum M | MMAR_2081 | - | 4e-11 | 30.65% (186) | transcriptional regulator |
| M. avium 104 | MAV_3499 | - | 3e-12 | 27.08% (192) | hypothetical protein MAV_3499 |
| M. smegmatis MC2 155 | MSMEG_5309 | - | 1e-74 | 62.96% (216) | hypothetical protein MSMEG_5309 |
| M. thermoresistible (build 8) | TH_1605 | - | 2e-70 | 64.59% (209) | conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_3263 | - | 2e-11 | 30.65% (186) | transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_2050 | - | 5e-68 | 60.29% (209) | putative transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2654c|M.bovis_AF2122/97 -------------------MGVS------VIIRSLQEPVGRRR-AVLRAL
Rv2621c|M.tuberculosis_H37Rv -------------------MGVS------VIIRSLQEPVGRRR-AVLRAL
MMAR_2081|M.marinum_M -------------------MGE----GR--IARMGGEPAGRRR-AVLRVL
MUL_3263|M.ulcerans_Agy99 -------------------MGE----GR--IARMGGEPAGRRR-AVLRVL
MAV_3499|M.avium_104 -------------------MGDPLTKSRKLLAPSAGESAGRRR-AVLRLL
Mflv_4298|M.gilvum_PYR-GCK -------------------MAVL---SSEAGQ-KPDRAAGRQRRRVLDLL
Mvan_2050|M.vanbaalenii_PYR-1 -------------------MAVQ---GRDTDK-ASDRAAGRQRQKVLGLL
TH_1605|M.thermoresistible__bu -------------------VSAP---RHDLDAPQSERAAGRQRRKVLDVV
MSMEG_5309|M.smegmatis_MC2_155 ----------------------------MTSAPPSDRSAGRQRKRVLEVV
MAB_0215|M.abscessus_ATCC_1997 ----------------------------MTGS----RGAGQQRARVLKVL
MLBr_00592|M.leprae_Br4923 MLNVHRGYPSLDRSPELRHTGVVKIRSELDAATTVAVSDGHTRRAVVRLL
*: * *: :
Mb2654c|M.bovis_AF2122/97 CASRVPMSIAAIAGKLGVHPNTVRFHLDNLVADGQVERVEPGR-GRPGR-
Rv2621c|M.tuberculosis_H37Rv CASRVPMSIAAIAGKLGVHPNTVRFHLDNLVADGQVERVEPGR-GRPGR-
MMAR_2081|M.marinum_M RALGHPLSIVAIAEHLGVHPNTVRFHLDGLVDDGQVERVQPSR-RGPGR-
MUL_3263|M.ulcerans_Agy99 RALGHPLSIVAIAEHLGVHPNTVRFHLDGLVDDGQVERVQPSR-RGPGR-
MAV_3499|M.avium_104 RASPEPMSIAGIADVLGVHPNTVRFHLDSLVADGQVEHVELDR-KGPGR-
Mflv_4298|M.gilvum_PYR-GCK READGPVDAQGVADLLKIHITTARFHLSTLETQGFVRR-AGVRKSGAGR-
Mvan_2050|M.vanbaalenii_PYR-1 RETKRPLDAQGVADTLQIHVTTARFHLSALESQGYVRRGRGPKAAGAGR-
TH_1605|M.thermoresistible__bu KRAAEPVDAQQVADALRIHVTTARFHLSTLEGQGAIRR-RRARNGGRGR-
MSMEG_5309|M.smegmatis_MC2_155 RRADGPVDAQQIADALQIHVTTARFHLGTLEEQGVIRRGGPGDKGRVGR-
MAB_0215|M.abscessus_ATCC_1997 QEAAAPVGAREVADSLGLHITTVRFHLKTLEEQGHIVRRGGTGEQRAGR-
MLBr_00592|M.leprae_Br4923 LESG-SITAGEISDRLGLSAAGVRRHLDVLIEMGDAESATAAPWQQVGRG
.: :: * : .* ** * * **
Mb2654c|M.bovis_AF2122/97 -PPLMFRAVRRTDSTGTRRYRLLAEILASGLAAERDSRAMALSAGRAWGR
Rv2621c|M.tuberculosis_H37Rv -PPLMFRAVRRTDSTGTRRYRLLAEILASGLAAERDSRAMALSAGRAWGR
MMAR_2081|M.marinum_M -PPLMFRAVQQMDRGGARHYQLLAEILSNGLAAESDSVDKALAAGRAWGE
MUL_3263|M.ulcerans_Agy99 -PPLMFRAVQQMDRGGARHYQLLAEILSNGLAAESDSVDKALAAGRAWGE
MAV_3499|M.avium_104 -PPLMFRAVRQMDRGGTRHYRLLAEILAKAFAAETDPAPKALAAGRAWGQ
Mflv_4298|M.gilvum_PYR-GCK -PKLTYEPAPRLDY--ADIVALFATHLG-GTAQEREARAQLIGADLAHRV
Mvan_2050|M.vanbaalenii_PYR-1 -PKLTYELAPRLDY--ADIVCLFATHLG-GTPQERESRARRIGADLAHRV
TH_1605|M.thermoresistible__bu -PRLTYEPTPRLDY--ADIVSLFATHLG-GTPVERERRATLIGADLARRV
MSMEG_5309|M.smegmatis_MC2_155 -PRLTYEIAPRLDY--ADIVALFARHLG-GTVEEREQRALRVGADLAHRV
MAB_0215|M.abscessus_ATCC_1997 -PSLSYAVAPRLDY--ADVVALFAVHLG-GTAAERESRAALVGADLAHRV
MLBr_00592|M.leprae_Br4923 RPAKHYRLTAVGRAKLGHAYDTLASAAMRQLREIGGEDAVRTFARQRIDA
* : . :* *
Mb2654c|M.bovis_AF2122/97 QLEA-----PPAGADTEETIDHLVAVLDDLGFAPERRASN-GRQQVGLR-
Rv2621c|M.tuberculosis_H37Rv QLEA-----PPAGADTEETIDHLVAVLDDLGFAPERRASN-GRQQVGLR-
MMAR_2081|M.marinum_M RLQA-----PTTAHSAEKSIDHLVGVLDGLGFAPERRVSD-GQQQVALR-
MUL_3263|M.ulcerans_Agy99 RLQA-----PTTAHSAEKSIDHLVGVLDGLGFAPERRVSD-GQQQVALR-
MAV_3499|M.avium_104 QLDAERRRLPRDAAGAEQAIAQLVDVLDELGFAPERRVID-GEQQVGLR-
Mflv_4298|M.gilvum_PYR-GCK RLAR--------PRAESSIGDLVVATLTELGFQVRSVLSSFGEVTVQLC-
Mvan_2050|M.vanbaalenii_PYR-1 RLAR--------PRDETSIADLVVATLTELGFQVRSLLSSFGEVTVQLC-
TH_1605|M.thermoresistible__bu RVAR--------RRDESTVADLVVETLGELGFQVRNVLTSFGEVTVQIC-
MSMEG_5309|M.smegmatis_MC2_155 RVAR--------RRDEESVVDMVVGTLTELGFQIRSTFTSFGTATVRIC-
MAB_0215|M.abscessus_ATCC_1997 NVAR--------RRTPLQVTDLVVETLGELGFTVRSTLMSFGRVTVQIC-
MLBr_00592|M.leprae_Br4923 ILADVEP--AGGGADVEAAANRVASALSDAGYVATTARVGGPIHGVEICQ
: :. .* *: . * :
Mb2654c|M.bovis_AF2122/97 -HCPFLELAETQAGVVCPVHLGIMRGALQTW-APVTVDRLDAFVEPD---
Rv2621c|M.tuberculosis_H37Rv -HCPFLELAETQAGVVCPVHLGIMRGALQTWGAPVTVDRLDAFVEPD---
MMAR_2081|M.marinum_M -HCPFLELAENRSGVVCPIHLGLMRGALESWGAPVTVERLDPFVEPD---
MUL_3263|M.ulcerans_Agy99 -HCPFLELAENRSGVVCPIHLGLMRGALESWGAPVTVERLDPFVEPD---
MAV_3499|M.avium_104 -HCPFLELAENSSNVVCPVHLGLMQGAMESWGAPVSVDRLDPFVEPD---
Mflv_4298|M.gilvum_PYR-GCK -TCPLAEVAQSAPEVVRGIQQGLIQEVIDLN-VDVIGGSYGVSVTPDPRG
Mvan_2050|M.vanbaalenii_PYR-1 -TCPLAEVAKAAPEVVRGIQQGMIQEVIDVN-AASIGGNYSVAVTPDPHA
TH_1605|M.thermoresistible__bu -TCPLAEVAETAPEVVRGIQQGLIQEVVDRN-AEVIGGRYRVAVTPDPRA
MSMEG_5309|M.smegmatis_MC2_155 -TCPLAEVAVDAPEVVRGVQQGLIQEVIDMN-ADAVGGRFTVSVRPDAKG
MAB_0215|M.abscessus_ATCC_1997 -SCPLAEIATTAPEVVRGIQRGLIQEVLDVN-ADAIGGQFQVTVSPDAGH
MLBr_00592|M.leprae_Br4923 HHCPVSHVAEEFP-ELCETEQQAIAEVLGTH-----VQRLAMIVNGD---
**. .:* . : . : .: * *
Mb2654c|M.bovis_AF2122/97 -LCLAHFTPLEGAIR---------------------
Rv2621c|M.tuberculosis_H37Rv -LCLAHFTPLEGAIR---------------------
MMAR_2081|M.marinum_M -LCLAHLS-PEGALS---------------------
MUL_3263|M.ulcerans_Agy99 -LCLAHLS-PEGALS---------------------
MAV_3499|M.avium_104 -LCLAHLT-LQGADR---------------------
Mflv_4298|M.gilvum_PYR-GCK GSCEVGLVLRPTRVAASEREGV--------------
Mvan_2050|M.vanbaalenii_PYR-1 GSCEIGLILRPARG----------------------
TH_1605|M.thermoresistible__bu GSCEVGLILRPGD-----------------------
MSMEG_5309|M.smegmatis_MC2_155 GSCEVSLVVSPVTRSGWCTGTTAANNCHGPTHLSQA
MAB_0215|M.abscessus_ATCC_1997 GDCTVNLALVPLGKESWPWT----------------
MLBr_00592|M.leprae_Br4923 CACTTHVPLMSVPSKT--------------------
* .